This article covers
What is a ribosome? A quick definition
A ribosome is a particle-like cell organelle made of RNA (ribonucleic acid) and ribosomal proteins that serve as the site for protein synthesis in the cell. Ribosomes consist of two major components: the small and large ribosomal subunits. Scientists like to call ribosomes, the macromolecular machines, to admire how exquisite the design of ribosomes is!
The ribosome reads the sequence of the messenger RNA (mRNA) and, using the genetic code, translates the sequence of RNA into a sequence of amino acids (a process called Translation).
[In this figure] An analogy for ribosomes in a factory.
Ribosomes work like machines to translate the code sequence of mRNA into a protein. Scientists like to call ribosomes, the molecular micro-machines, to admire how exquisite the ribosomes’ design is!
Ribosomes structure
A ribosome is a complicated but elegant “micro-machine” for producing proteins. Ribosomes are made up of ribosomal proteins and ribosomal RNA (rRNA). In prokaryotes, ribosomes consist of roughly 40 percent protein and 60 percent rRNA. A eukaryotic ribosome comprises three or four rRNA molecules and about 80 different proteins. Its’ molecular mass is about 4,200,000 Da. About two-thirds of this mass is ribosomal RNA, and one-third of that is different ribosomal proteins. Ribosomes are not membrane-bound organelles.
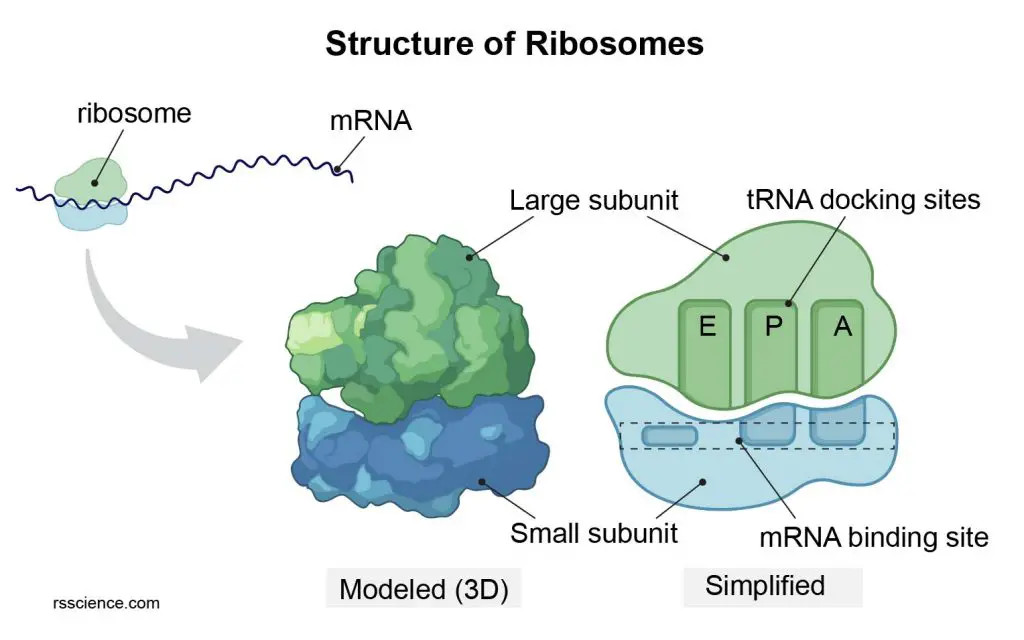
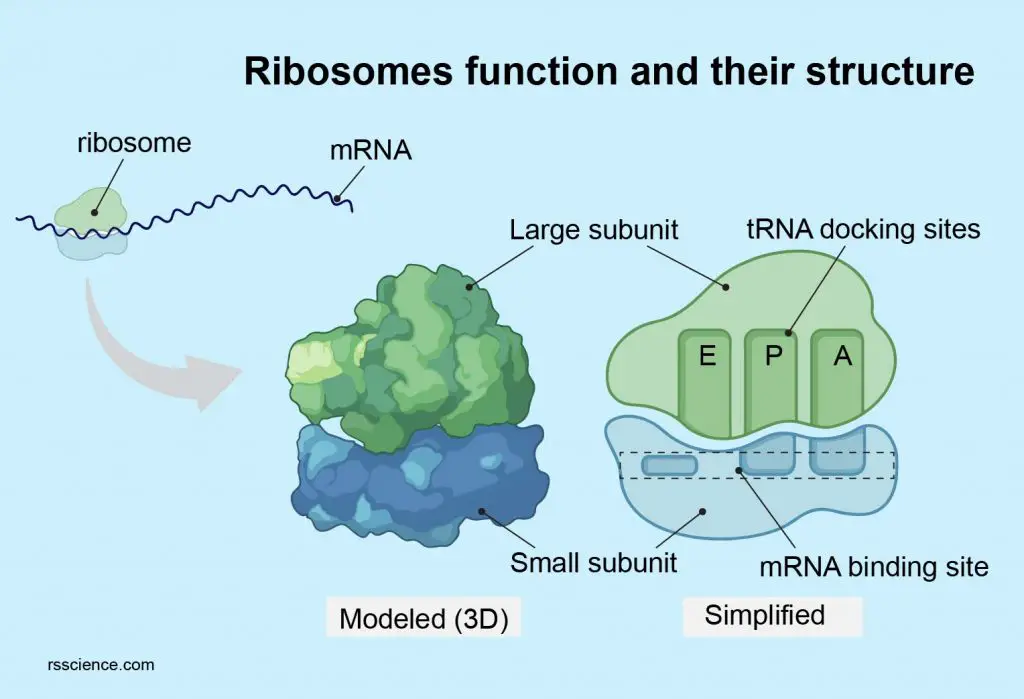
[In this figure] Diagram of ribosome structure showing the large and small subunits.
Each ribosome comprises two subunits, a larger subunit, and a smaller subunit; both are RNA-protein complexes. The larger subunit has catalytic activity, while the smaller subunit function as a decoding machine.
The name of subunits is based on their sedimentation rate, meaning how fast they can be settled down when a mixture of cell lysis was span in a centrifuge. This rate is measured in Svedberg units (S) rather than size. This is why subunits’ names do not add up: for example, bacterial 70S ribosomes are made of a large 50S and a small 30S subunits, while human 80S ribosomes contain 60S and 40S subunits.
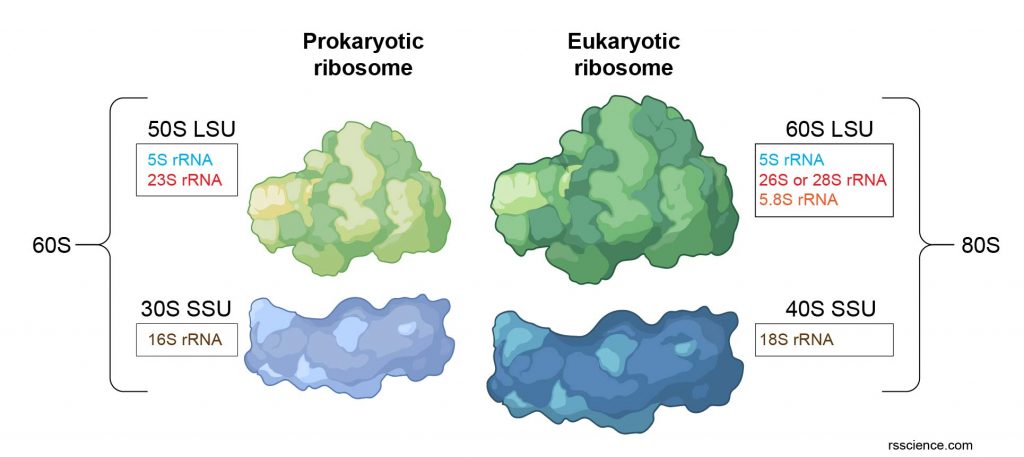
[In this figure] The composition of prokaryotic ribosome rRNA and eukaryotic ribosome rRNA. LSU is a large subunit and SSU is a small subunit.
Ribosomal ribonucleic acid (rRNA) is the primary component of ribosomes and carries our protein synthesis like an enzyme; therefore also called ribozyme. Unlike messenger RNA, rRNA does not carry genetic information. Thus, they are non-coding RNA. Ribosomal RNA is transcribed from ribosomal DNA (rDNA).
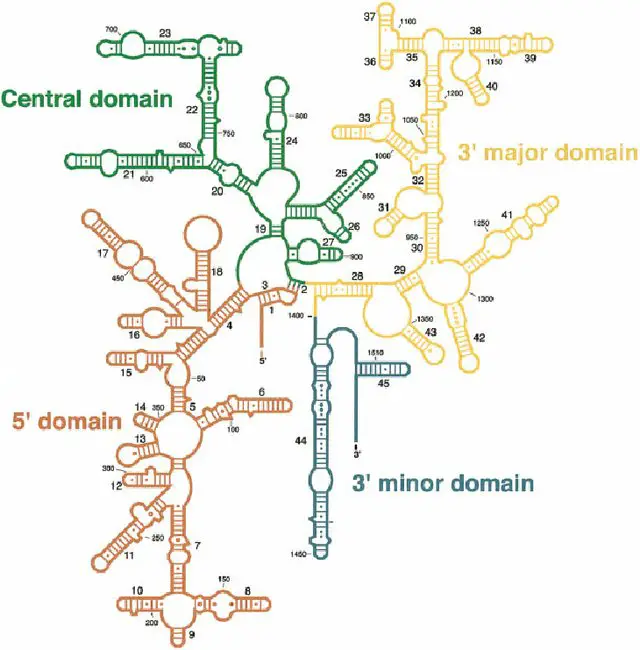
[In this figure] The secondary structure of rRNA.
rRNA is not a linear RNA molecule. In fact, it folds into a specific 3-D structure with stems and loops. Stems are short double-stranded RNAs formed by base-pair complementarity (A=U, G≡C). Loops are unpaired single-stranded RNAs. This image is a secondary structure of 16S rRNA. The tertiary structure of rRNA will fold this structure up among a complex of proteins. The entire 16S rRNA gene sequence is approximately 1500 base pairs (bp).
Photo credit: research gate
Ribosome function – protein translation

[In this figure] The ribosome works like a machine to translate the code sequence of mRNA into a protein.
Ribosomes are the molecular machines responsible for protein synthesis (called Translation). At the molecular level, ribosome functions like a decrypting machine.
DNA transcribes to messenger RNA (mRNA), which is exported from the nucleus to the cytoplasm. The mRNA molecule is like genetic codes written on a long thread of paper. The ribosome finds the right end of the mRNA thread and starts to read the codes.
The ribosome uses the code sequence for determining the correct order of amino acids to generate the corresponding protein. During the process, the large subunit sits on top of the small subunit, with an RNA template sandwiched between the two. (A ribosome looks like a hamburger with a puffy bun on top, an RNA “patty” threading through it.)
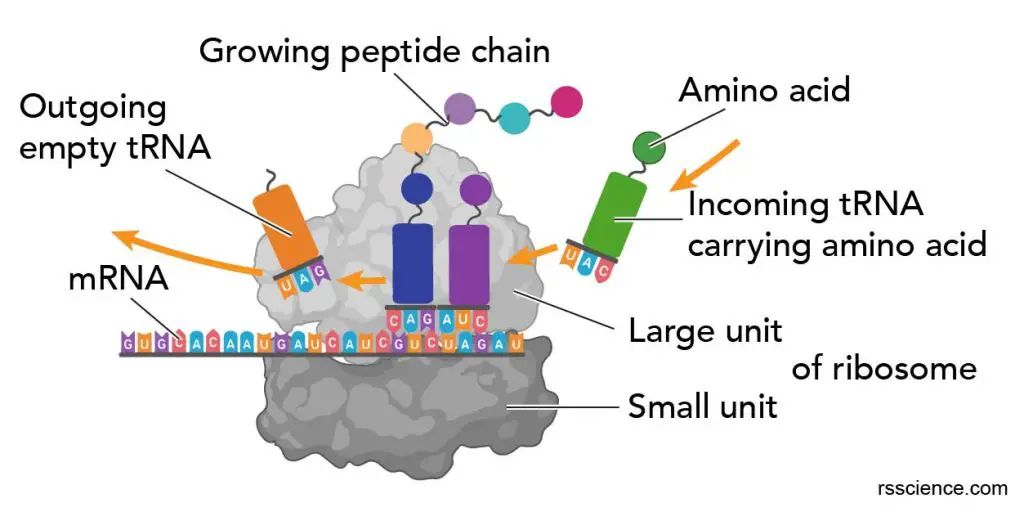
Three tRNA binding sites – A, P, and E sites
The ribosome has three binding sites for tRNA: A, P, and E sites. A and P sites span both the ribosome subunits with a larger part residing in the ribosome large subunit and a smaller part in the smaller subunit. E site (the exit site) resides in the large ribosome subunit.
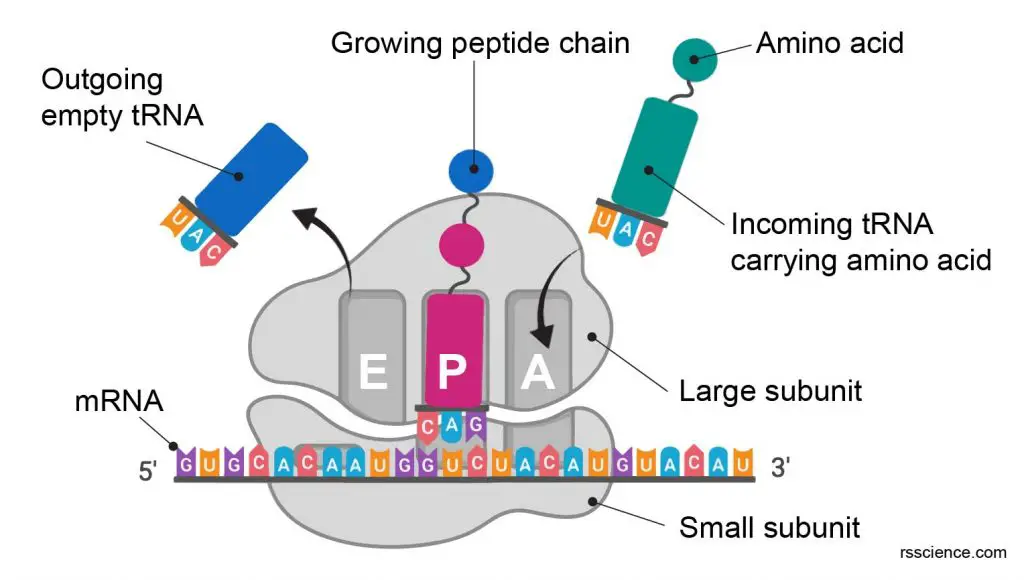
[In this figure] The RNA binding sites on a ribosome.
The ribosome contains 3 RNA binding sites: A, P, and E sites for protein translation.
The RNA binding sites, positions, and their functions in a ribosome:
| Binding Site | mRNA strand entry site | Biological term | Main processes |
| A site | 1st | Aminoacyl | Admission of the codon of mRNA & “charged” tRNA. Checking and decoding and start of “handing over” one amino acid molecule |
| P site | 2nd | Peptidyl | Peptide synthesis, consolidation, elongation and transfer of peptide chain to site A |
| E site | 3rd | Exit | Preparation of “uncharged” tRNA for exit |
tRNA
The transfer RNA (tRNA) is one type of RNA molecule. Its job is to carry the amino acid that matches the mRNA codon to the ribosome.
The tRNA contains a three-letter code on one side and carries a specific amino acid on the other side. The code on tRNA (called an anti-codon) must match the three-letter code (called a codon) on the mRNA already in the ribosome.
The particular amino acid that tRNA carries is determined by a three-letter anti-codon it bears. For example, if the three-letter code is AUG on mRNA (Adenine, Uridine, Guanine), the tRNA that carried Methionine (Met or M) will be selected and recruited to the ribosome. This is an essential part of the translation process, and it is surprising how few “errors of translation” occur.
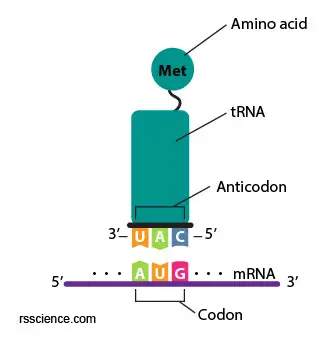
[In this figure] A anticodon UAG on the tRNA matches the AUG on the mRNA (complimentary) and brings the right amino acid to the ribosomes.
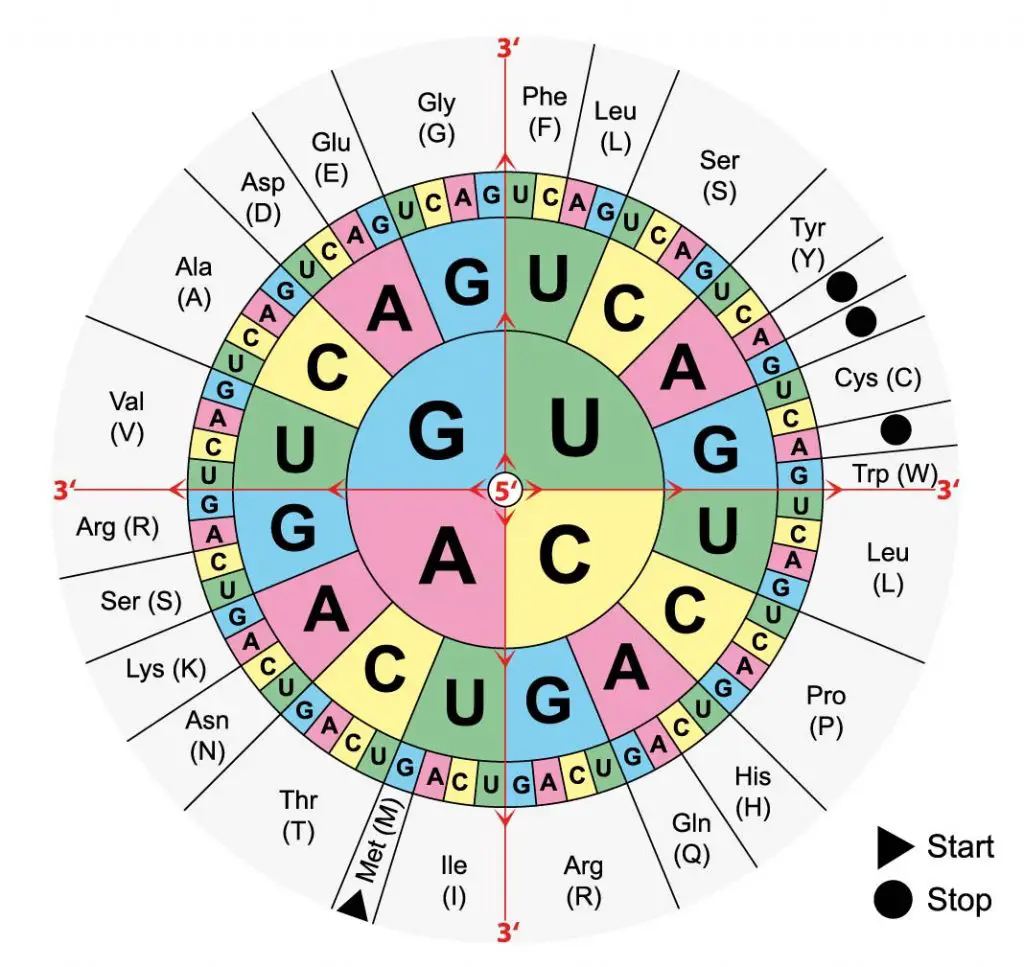
[In this figure] The standard mRNA codon table is organized in a wheel.
Read the first letter from the center. For example, UCG is Serine (Ser or S).
Protein translation process – 3 steps
The protein translation by a ribosome consists of three stages: (1) Initiation, (2) Elongation, and (3) Termination.
Initiation – the ribosome assembles around the target mRNA.
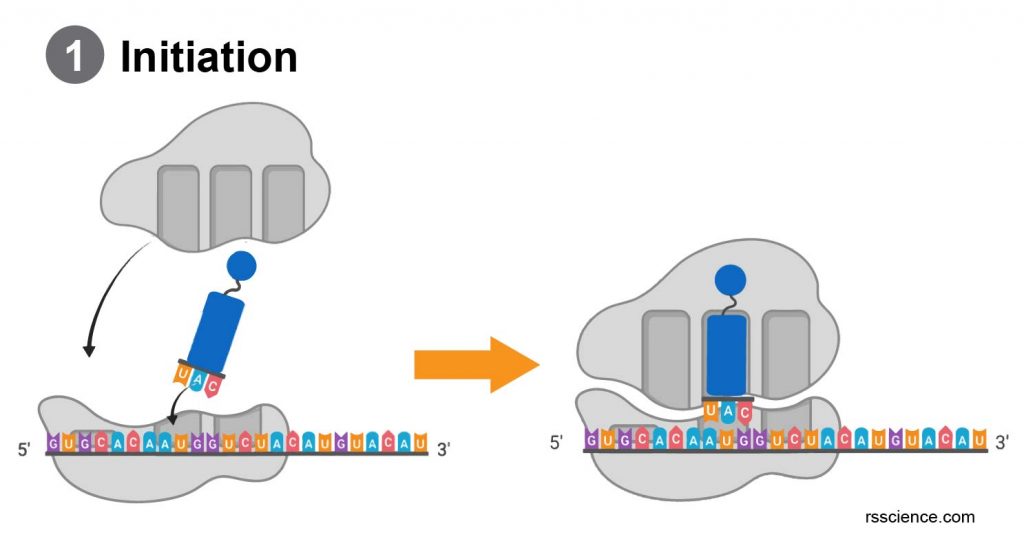
A small ribosome subunit links onto the “start-end” of an mRNA strand. “Initiator tRNA” also enters the small subunit and binds to the start codon (most commonly, AUG). Then, a ribosome large subunit joins this complex. The initiator tRNA will be at the P site.
Elongation
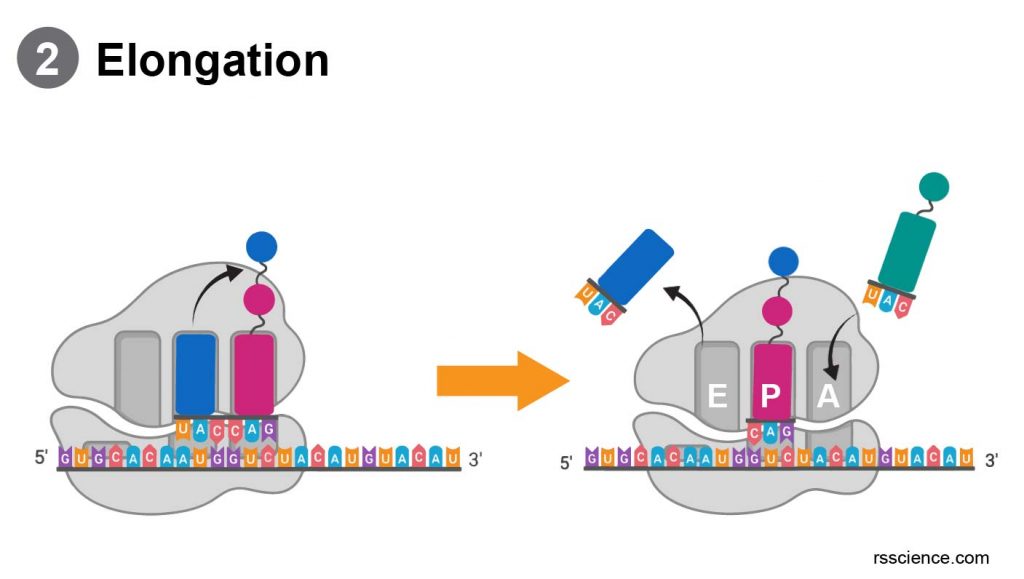
During the elongation process, the tRNA carries an amino acid to the ribosomes (A site). “Charged” tRNA means this tRNA carrying one amino acid residue. Only the right tRNA can enter the ribosome and pair with the code on mRNA. Once the tRNA and mRNA match, the previous tRNA (in the P site) will donate its growing protein chain onto this amino acid (still in A site). Then, both tRNAs move one site forward (A to P; P to E). “Uncharged” or empty tRNA will prepare to exit from the E site.
The next charged tRNA will enter the available A site. mRNA will also slide to the next codon to initiate the next translational cycle. After all the codes on the mRNA molecule successfully translate into the corresponding amino acids, the protein is synthesized.
Termination
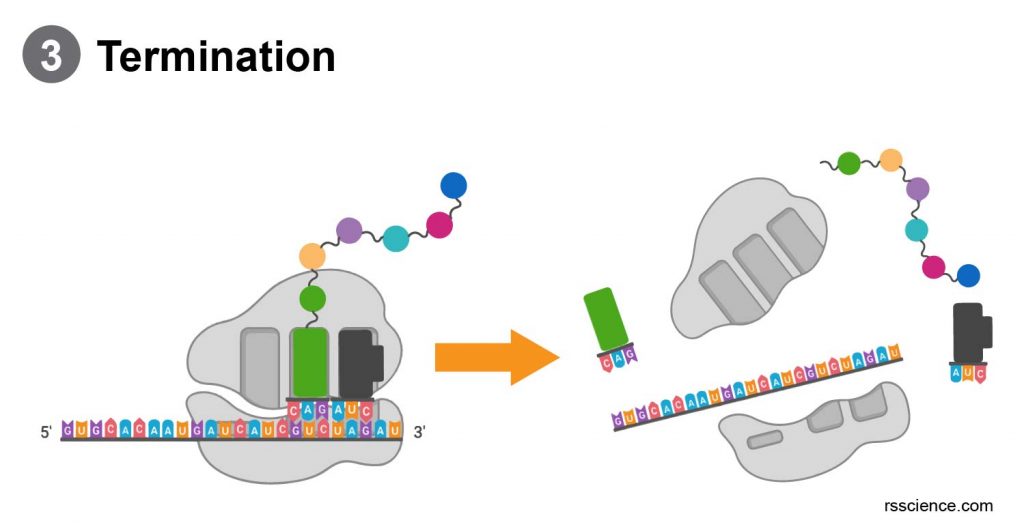
When the ribosome reaches the end of the mRNA strand (labeled by a stop codon), the ribosome releases the polypeptide. The two ribosome subunits disengage, separate, and are re-used or broken down.

[In this animation] A ribosome translating a protein.
How fast the ribosomes translate?
Ribosomes can translate at a rate of 200 amino acids per minute. Small proteins can therefore be made fairly quickly, but two to three hours are needed for larger proteins such as the massive 30,000 amino acid muscle protein titin.
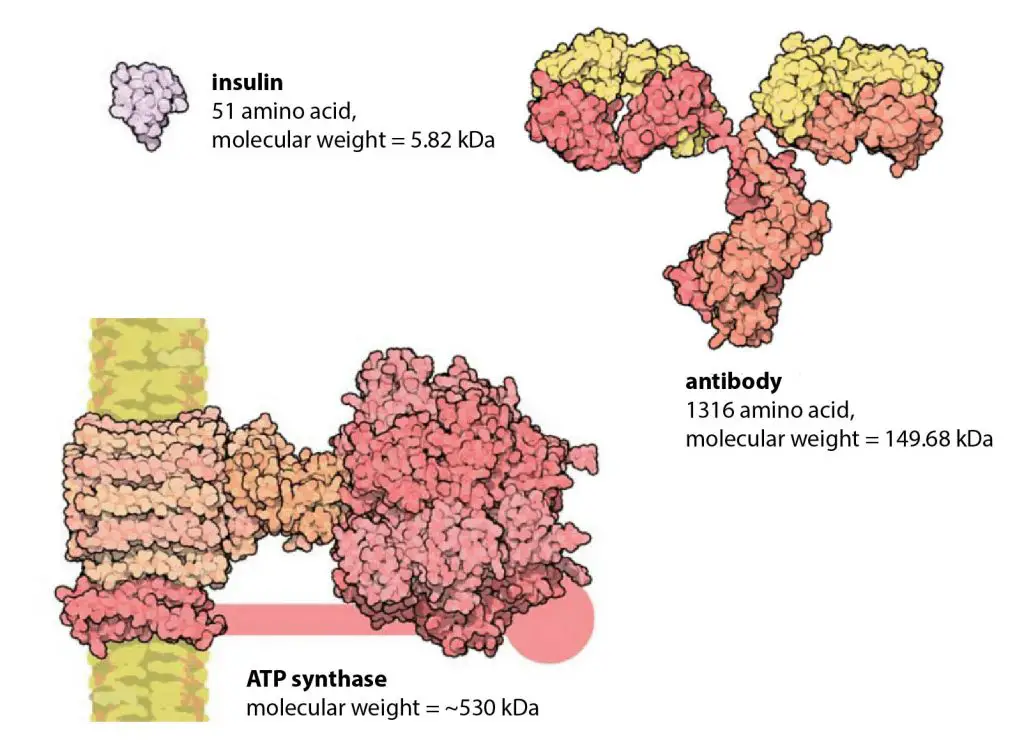
[In this figure] How big is the “average” protein? The median protein length in human cells is around 375 amino acids. All the proteins in the figure are shown on the same scale to give an impression of their relative sizes.
Photo credit: modified from Cell biology by the numbers
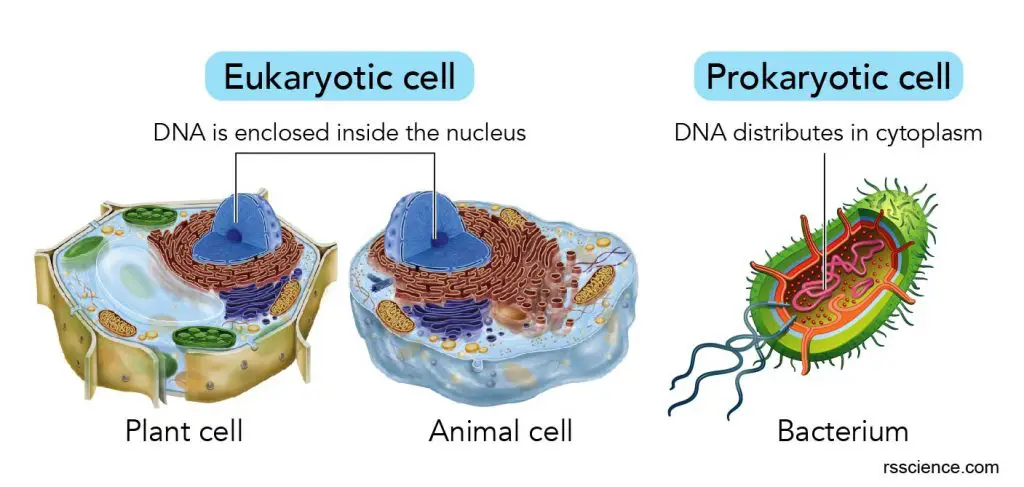
Do prokaryotes have ribosomes?
Yes, both prokaryotes (bacteria and archaea) and eukaryotes (animals, plants, fungi) have ribosomes. Ribosomes in prokaryotes use a slightly different process to produce proteins than ribosomes do in eukaryotes.
Fortunately, this difference presents a window of opportunity for attack by antibiotic drugs such as streptomycin. Unfortunately, some bacterial toxins and the poliovirus also use this gap to attack the human translation mechanism.
Ribosomes are also found in mitochondria and chloroplasts. Ribosomes found in prokaryotes are generally smaller than those in eukaryotes. Ribosomes in mitochondria and chloroplasts are similar in size to those in bacteria.
| Eukaryote | Prokaryotes | Mitochondrion | Chloroplast | |
| other names | cytoplasmic ribosome | cytoplasmic ribosome | mitoribosomes | plastoribosomes |
| location | free or rough ER | free in cytoplasm | free in matrix | free in stroma |
| size | 25-30 nm | 20 nm | 20 nm | 20 nm |
| rRNA/protein ratio | 50:50 | 65:35 | * | * |
| whole unit | 80S** | 70S | 55S | 78S |
| subunit | 40S and 60S | 30S and 50S | 28S and 39S | 33S and 50S |
| rRNA molecules per ribosome | 4 | 3 | 2 or 3 | 3 |
| rRNA | 28, 15, 5.8, 5S | 23, 16, 5S | 16, 12S |
* The exact ratio is undefined. Generally, ribosomes of mitochondria and chloroplasts are closer to the prokaryote’s ribosome.
** The unit to measure the ribosomal subunits and rRNA fragments is the Svedberg unit. It measures the rate of sedimentation in centrifugation rather than size. This is why fragment names do not add up.
The discovery of ribosomes
The small particles that are known as ribosomes were first described in 1955 by cell biologist George E. Palade, who found them using an electron microscope. He noticed that these particles are frequently associated with the endoplasmic reticulum in eukaryotic cells. The word ribosome is made from taking “ribo” from ribonucleic acid and adding it to “soma,” the Latin word for body.
The Nobel Prize in Physiology or Medicine in 1974 was awarded to the discovery of the ribosome (Albert Claude, Christian de Duve, and George Emil Palade). In 2009, the Nobel Prize in Chemistry was awarded to Venkatraman Ramakrishnan, Thomas A. Steitz, and Ada E. Yonath, who mapped ribosome structure and mechanism down to the level of individual atoms using a technique called X-ray crystallography.

[In this figure] The Nobel Prize in Physiology or Medicine 1974 for the discovery of the ribosome.
Photo credit: The Nobel Prize.

[In this figure] Atomic structure of the bacterial ribosome’s 30S subunit.
This work was done by a Nobel prize winner, Venkatraman Ramakrishnan.
Photo credit: Nature
Where are ribosomes located inside a cell?
Ribosomes can function in a “free” form in the cytoplasm, called free ribosomes. However, they can also “settle” on the endoplasmic reticulum (ER) to form “rough endoplasmic reticulum (RER).” Ribosomes in close association with the endoplasmic reticulum can facilitate the further processing of newly made proteins.
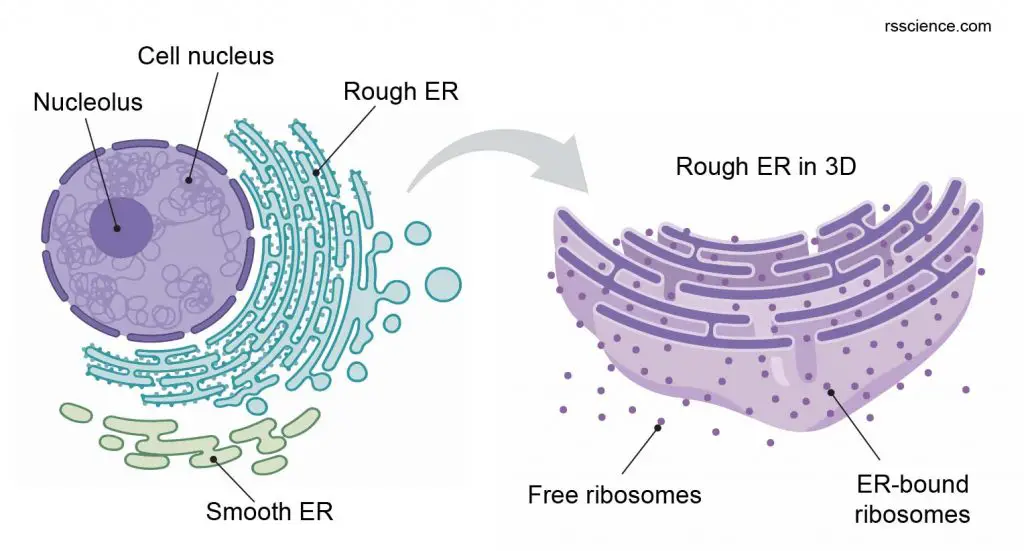
[In this figure] Ribosomes are the places where proteins were synthesized in our cells. Ribosomes can be found to be free-floating in the cytosol or associated with rough ER.
How many ribosomes in a cell?
There are about 10 billion protein molecules in a mammalian cell, and ribosomes produce most of them. A rapidly growing mammalian cell can contain about 10 million ribosomes. This number is highly dependent on the cell types and the status of cells.
Ribosomes are particularly abundant in cells that synthesize large amounts of protein. For example, the pancreas is responsible for creating several digestive enzymes, and the cells that produce these enzymes contain many ribosomes. Another example is the immature red blood cells (reticulocytes). Before they become mature, they have to synthesize a lot of hemoglobin. During this differentiation process, ribosomes are particularly abundant in immature reticulocytes.
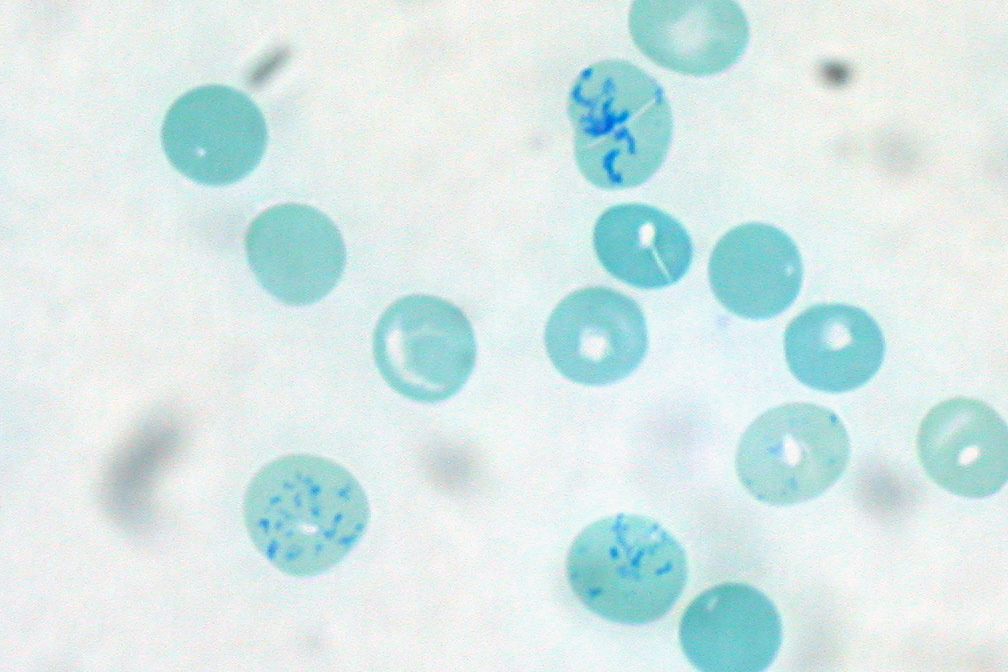
[In this figure] The reticulocytes contain dark blue dots and curved linear structures (reticulum) in the cytoplasm. They are called reticulocytes because the mesh-like network of ribosomal RNA becomes visible under a microscope with certain stains.
Photo credit: Ed Uthman, MD
In the bacterium, a single cell of Escherichia coli (E. coli) contains as many as 15,000 ribosomes and this accounts for one-quarter of the cell’s total mass.
Where are ribosomes made of?
The proteins and nucleic acids that form the ribosome subunits are made in the nucleolus (in the nuclei) and exported to the cytoplasm through nuclear pores. The two subunits stay apart until required for use. Ribosomes are not static organelles. When specific protein production is finished, two subunits separate and are usually broken down. The intact ribosomes are temporary existence.
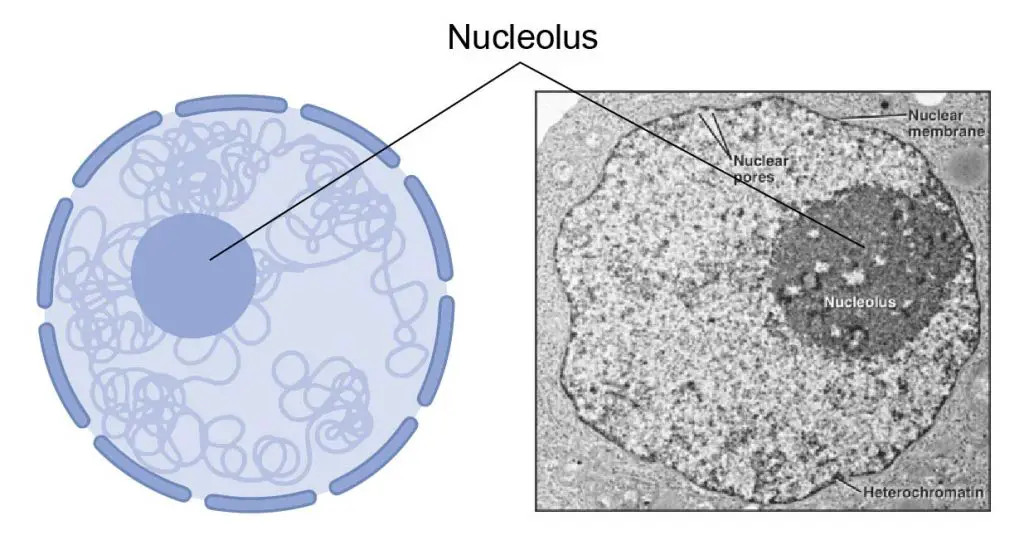
[In this figure] The main function of the nucleolus is to produce ribosomes and synthesize ribosomal RNA (rRNA).
What is a Polysome?
A polysome (polyribosome or ergasome) is a group of ribosomes bound to an mRNA molecule like “beads” on a “thread.” Multiple ribosomes move along the coding region of mRNA and translate several proteins like a production line.
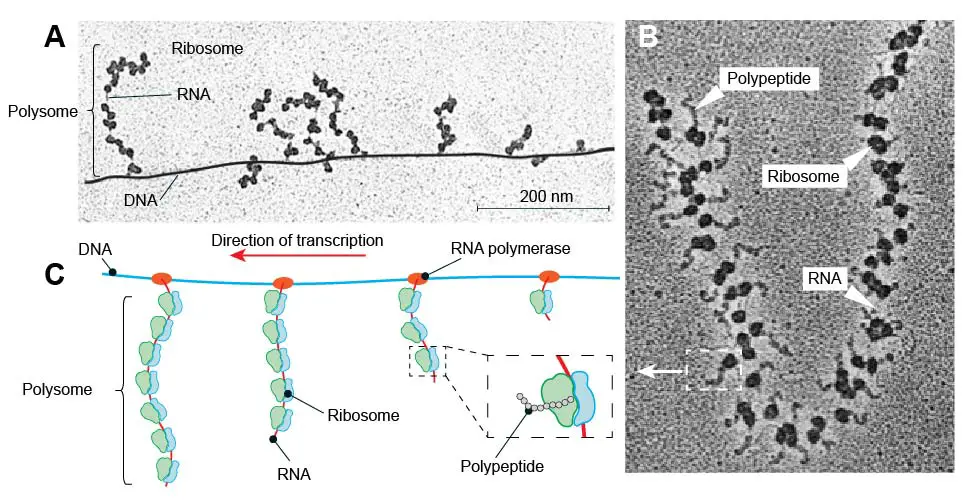
[In this figure] (A) TEM image of prokaryotic polysomes on DNA. (B) TEM image of a polysome with many ribosomes translating polypeptide chains. (C) Schematic diagram of prokaryotic polysomes in (A).
Prokaryotes lack a nucleus, so their mRNAs are transcribed in the cytoplasm and can be translated by ribosomes immediately. As a result, you can even see several polysomes attached to a single DNA molecule in bacterium cells. This can tell us the direction of transcription because the polysome located at the 3’-end of DNA should be longer than those at the 5’-end.
What does co-translational translocation mean?
In eukaryotes, translation happens when ribosomes bind to the rough endoplasmic reticulum (ER). These ribosomes bind to the rough ER’s outer membrane, and the new protein is synthesized across the membrane of ER. The newly created proteins can be stored inside the ER for future vesicle transport and secretion outside the cell or immediately secreted. This process is called co-translational translocation.
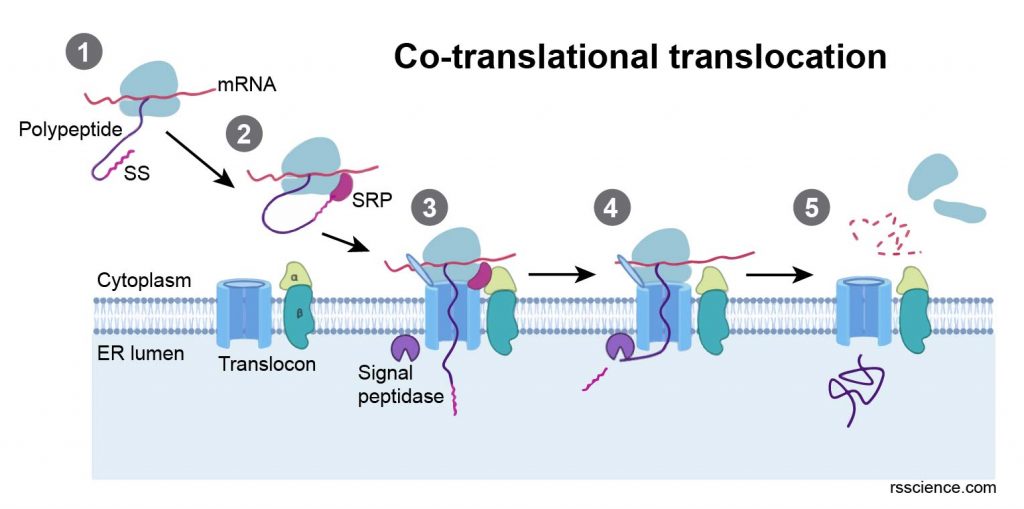
[In this figure] The process of co-translational translocation: (1) newly translated polypeptide presents a signal sequence (SS), (2) the signal recognition particle (SRP) recognizes SS, (3) the SRP receptor recognizes SRP, allowing the ribosome to bind the translocon and the polypeptide to translocate into the ER lumen, (4) translation resumes and the SS is cleaved by a signal peptidase, (5) translation completes in the ER.
What is the ribosome binding site?
A ribosome binding site (RBS) is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of protein translation. Mostly, RBS refers to bacterial sequences, while an internal ribosome entry site (IRES) refers to eukaryotic cells. Ribosome recruitment in eukaryotes is generally mediated by the 5′ cap present on eukaryotic mRNAs.
What is the ribosome profiling?
Ribosome profiling or Ribo-Seq is a sequencing technique that can determine which mRNAs are being actively translated (known as a translatome). Ribosome profiling can be used to identify translated mRNA regions (i.e., starting codon) and measuring protein synthesis.
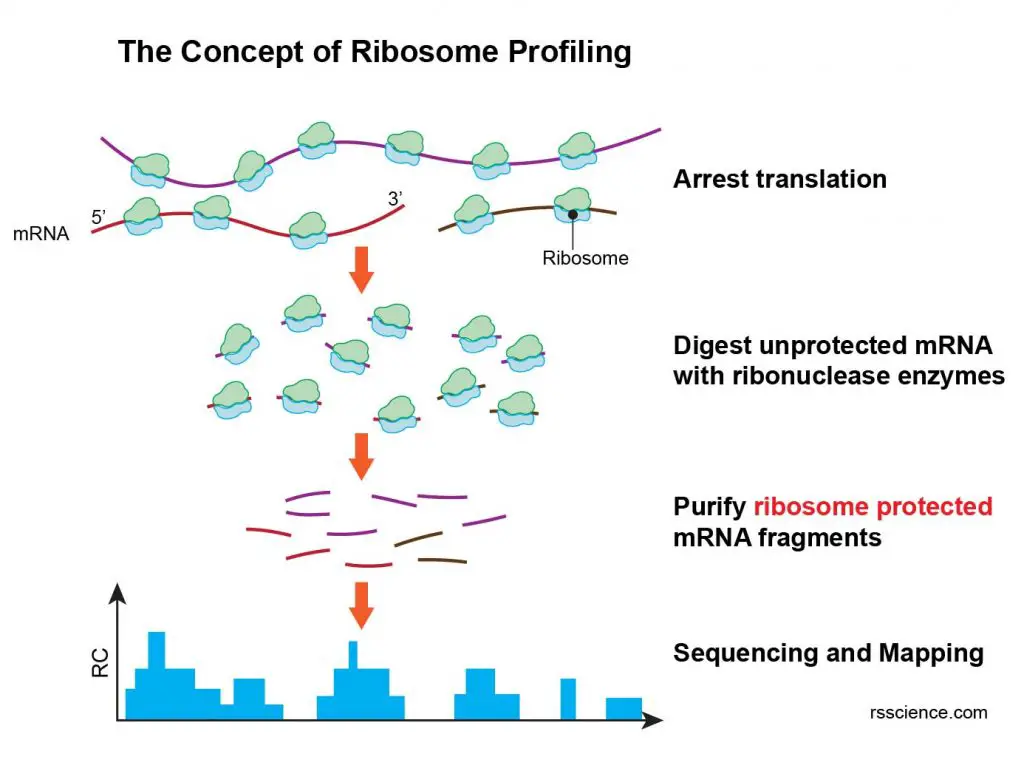
[In this figure] The concept of ribosome profiling.
Ribosomes actively translated on mRNA are arrested, then exposed mRNA outside of the ribosome is digested. Protected mRNA footprints are then sequenced by next generation sequencing and mapped to the genome, creating for each gene its read count (RC) profile. Therefore, ribosome profiling provides a snapshot of protein production in cells at a specific time point.
Ribosomes under a microscope
Why ribosomes can not be seen through a regular light microscope?
Some cell organelles, including ribosomes, ERs, lysosomes, centrioles, and Golgi bodies, cannot be seen with light microscopes because these microscopes cannot achieve a magnification high enough to see these relatively tiny organelles.
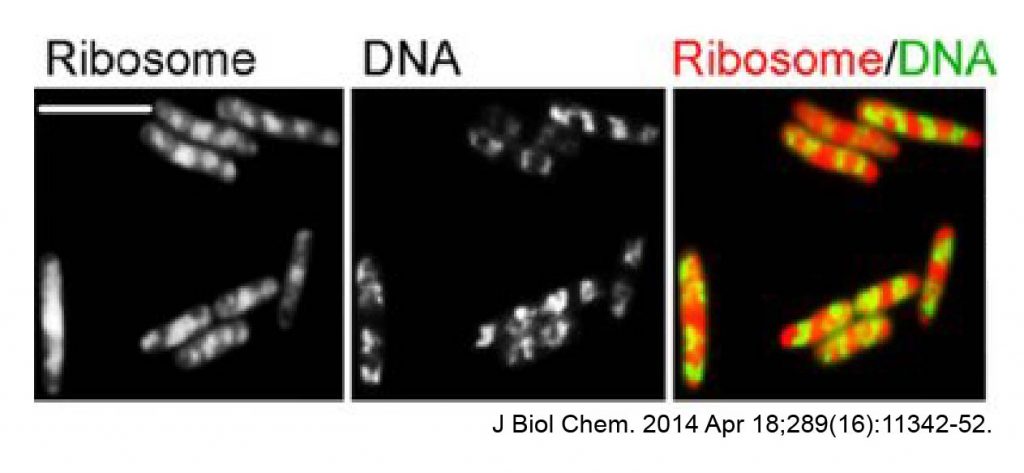
[In this figure] Ribosomes under a fluorescence microscope.
Recently, scientists applied super-resolution fluorescence microscopes to study ribosomes in bacteria. However, the resolution is still very limited.
Ribosomes under an electron microscope
A transmission electron microscope (TEM) is the best tool to observe ribosomes.
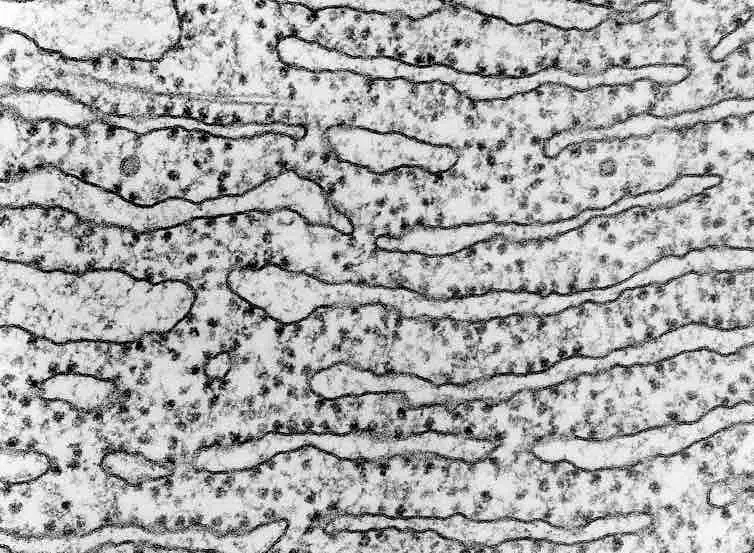
[In this figure] An electron microscope image of the rough endoplasmic reticulum. The dark spots are ribosomes.
Summary
- Ribosomes synthesize nearly all the proteins required by cells.
- Ribosomes are found free-floating in the cytoplasm or attached to the rough endoplasmic reticulum.
- Ribosomes consist of two parts: small and large subunits. Both subunits are ribonucleoprotein (RNA-protein) complexes.
- Ribosomes are produced in the nucleolus.
- Ribosomes translate information encoded in mRNA and link specific amino acids to form polypeptides. This process is called protein translation.
- Specific interactions between mRNA, ribosome, and tRNA take place at A, P, and E sites on ribosomal large and small subunits.
- Three stages in the translation process: (1) Initiation, (2) elongation, and (3) termination.
- Ribosomes are too small to be seen by a light microscope. Instead, a transmission electron microscope (TEM) is required to view ribosomes and rough endoplasmic reticulum.
References
“Ribosome”
“Ribosome” by British Society for Cell Biology
“Nucleus and ribosomes”
“Structure and Function of the Eukaryotic Ribosome” by Jennifer A Doudna and Virginia L Rath
“Ribosome database project”

Pingback: What type of RNA carries amino acids to the ribosome? – Dr. Biology Questions and Answers
Pingback: What is a ribosome? – Dr. Biology Questions and Answers
Pingback: What does a ribosome do? – Dr. Biology Questions and Answers
Pingback: Which of these organelles manufactures proteins bound for secretion out of the cell? – Dr. Biology Questions and Answers