This article covers
What are chromosomes? A quick overview
Chromosomes are bundles of tightly coiled DNA molecules. In eukaryotic cells, chromosomes are stored in the nuclei of cells. We have 23 pairs of chromosomes (1-22, X, and Y), and the number will double right before the cell division.
When cells are ready for division, their long DNA threads will be compressed into chromosomes. Packed DNA make the proper separation of equal genetic materials into daughter cells easier.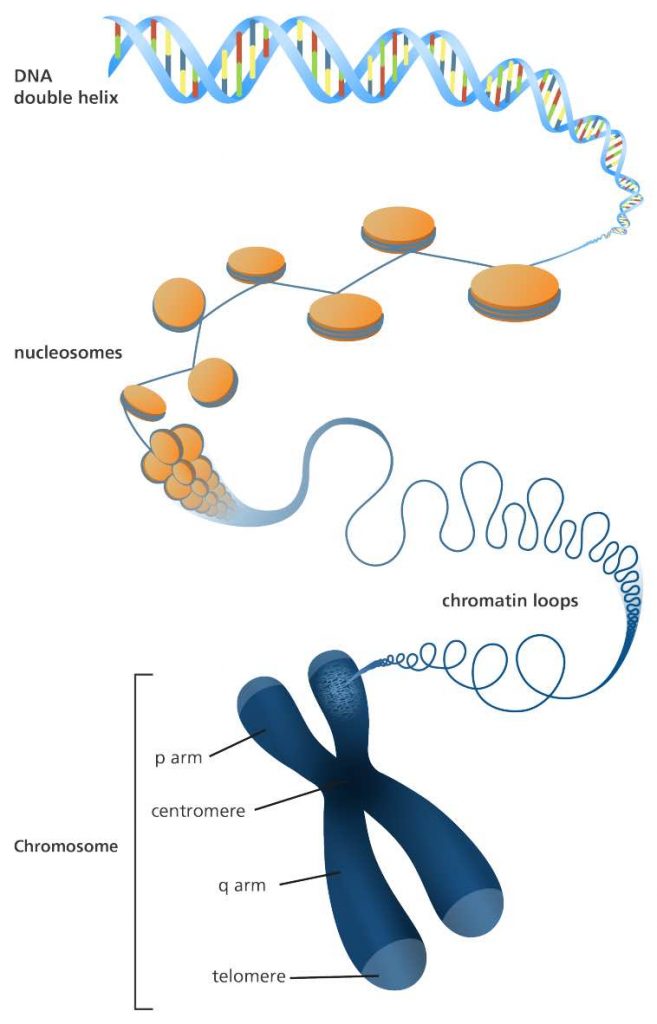
[In this image] A illustration of how a long DNA molecule is packaged into a short chromosome. Each chromosome is made up of DNA that are tightly coiled many times around proteins called histones. The material that contains DNA and proteins is called chromatin.
Image credit: Genome Research Limited
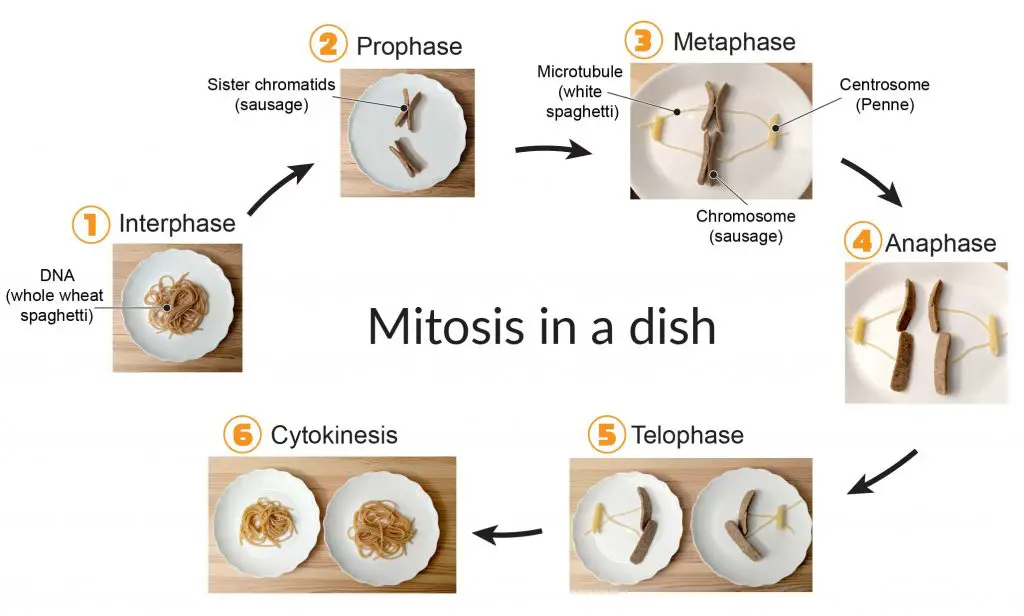
[In this image] Would you like to learn the cell biology hand-on? See how you can create a fun food model of mitosis using sausages as chromosomes and spaghetti as microtubules!
How cells store their genetic information
Our genes are encrypted books carrying the secrets of life. The language of these gene books is written in the nucleotide base pairs (A, T, G, C). The papers of these books are made of DNA molecules.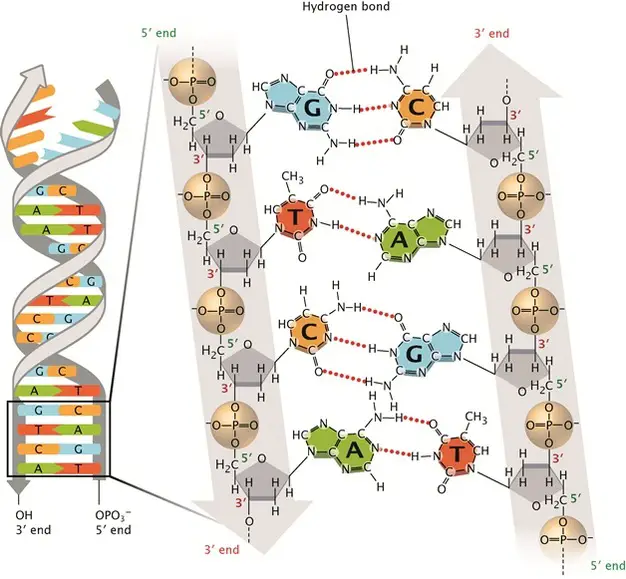
[In this image] DNA double-stranded helix.
The DNA alphabets contain only four letters, which are four nitrogenous base options: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). Each base can only pair with one other, A with T and C with G. This is called the DNA complementary base pairing rule. By this rule, two complementary DNA strands pair together and form a double-stranded DNA helix.
All of our genes are stored in the cell nuclei, located on 46 pieces of long DNA threads. The entire set of an organism’s genetic materials is called “genome”. The science to study the genomes is called “genomics”.
Don’t mess up with DNA
Except for a dividing cell, DNA molecules in the nucleus are in the “open” form so that each gene is accessible to be transcribed into mRNA.
If you stretch all the DNA from a single cell into a linear thread, it can be as long as 2-3 meters. It is incredible how the cell can pack entire DNA into a tiny nucleus (1 million times compacted, the diameter of a nucleus is less than 2-3 micrometers; one micrometer = 0.000001 meters).
All the genetic materials will be duplicated when a cell is ready for division. One copy of the entire genome must distribute to each daughter cell properly. Otherwise, the cell division fails and may result in diseases like cancer.

[In this figure] Tangled earphones are one of the most annoying things in our life. In our cell, the DNA threads are even longer than the earphone’s wire.
As you can imagine, it is almost impossible to separate two sets of DNA molecules when they are so long and tangled. To solve this problem, each DNA thread is organized into a much compact structure, called “chromosome”. In human cells, 46 pieces (or 23 pairs) of long DNA threads are compressed into 23 pairs of chromosomes (1-22, and X or Y).
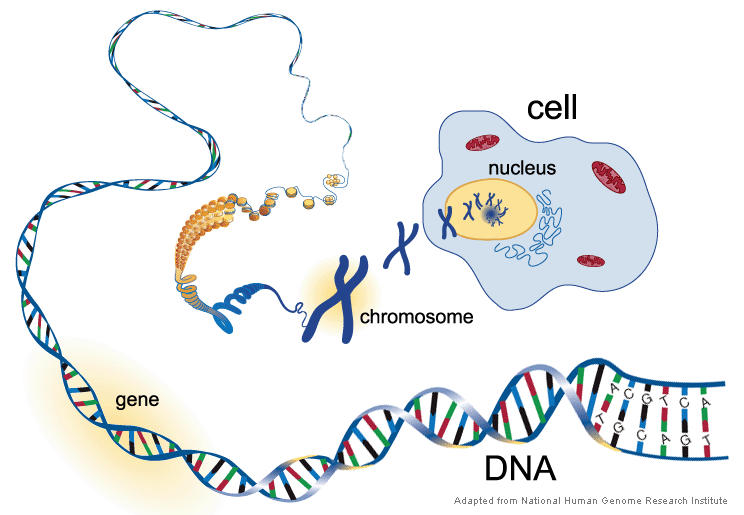
[In this figure] In order to handle the long DNA molecules, our cells pack DNA threads into many compact structures, called “chromosomes”.
Click here to jump to the section “How to Pack DNA into chromosomes”.
Can we see DNA and chromosomes under a microscope?
DNA threads are not visible in the cell’s nucleus – not even under a microscope. This is because a DNA helix is super thin, measured only 2.2-2.6 nm wide. However, when the cell is preparing for cell division, the DNA packs into chromosomes and becomes more tightly condensed; therefore it is visible under a light microscope.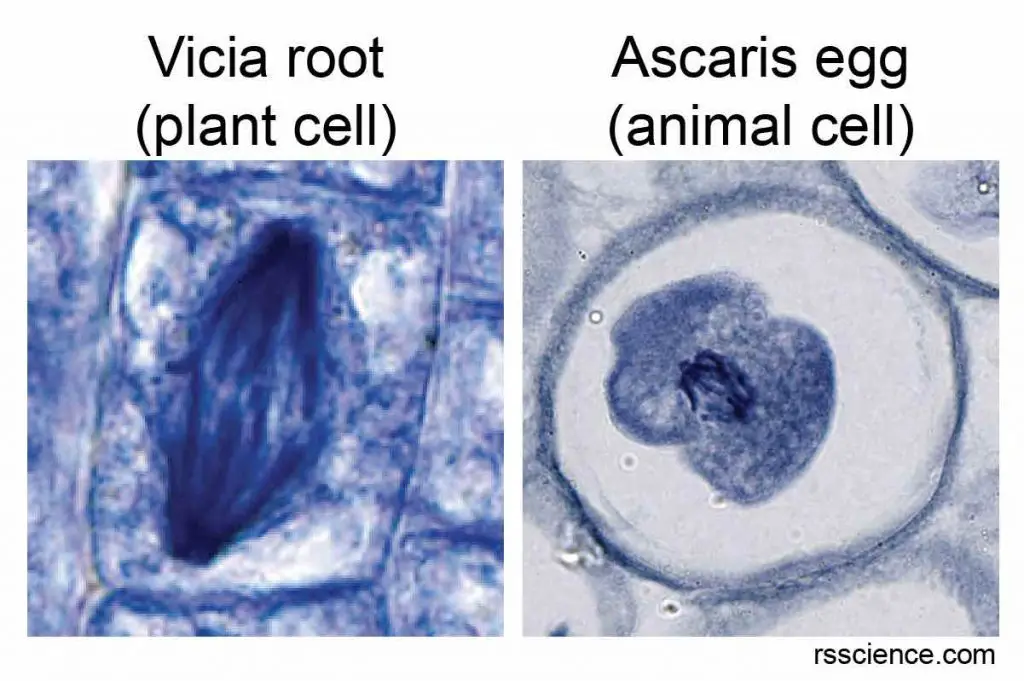
[In this image] Two examples of chromosomes inside dividing cells.
Some cells, such as root cells near the root’s tip and cells in animal eggs, have bigger nuclei and are actively dividing. You have a good chance to see their chromosomes under a microscope.
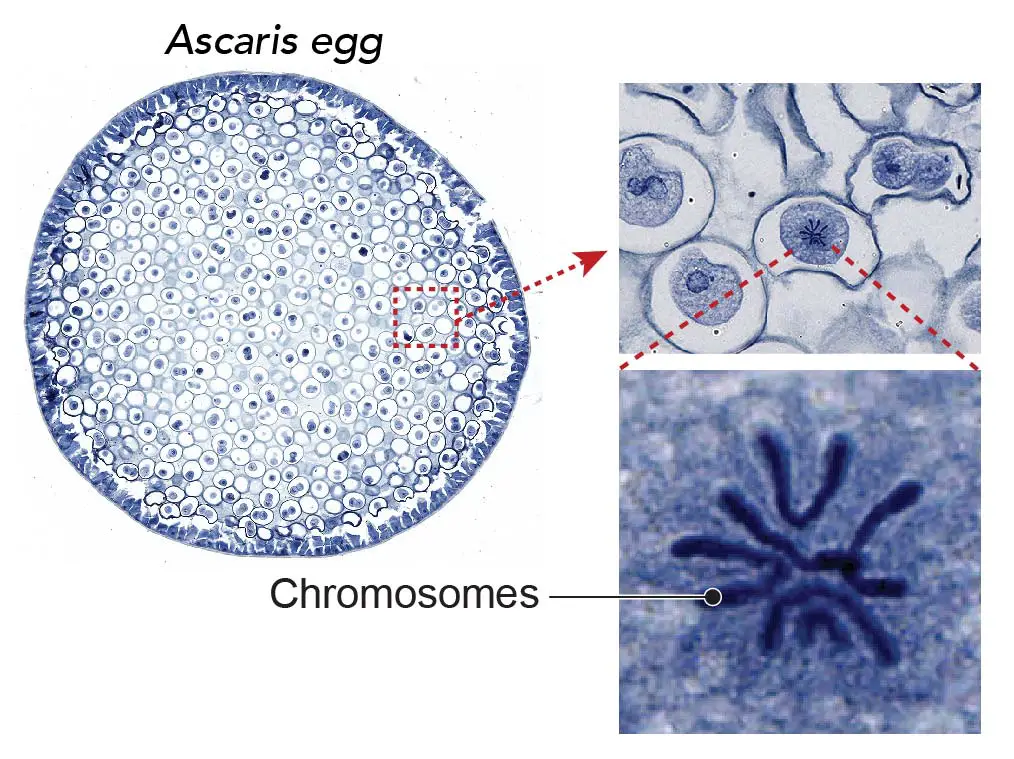
[In this image] Chromosomes are visible during cell division.
The chromosomes become visible under the light microscope with 40x-100x objective lens. Methylene blue stains the DNA contents of chromosomes with dark blue color. This specimen is Ascaris (parasitic nematode worm) eggs that contain active mitotic cells. This specimen is included in Rs’ Science 25 Premade slide set.
The structure of chromosomes
Each chromosome has a constriction point called the centromere, which divides the chromosome into two sections, or “arms.” The short arm of the chromosome is labeled the “p arm.” The long arm of the chromosome is labeled the “q arm.” The ends of the chromosome are called telomeres.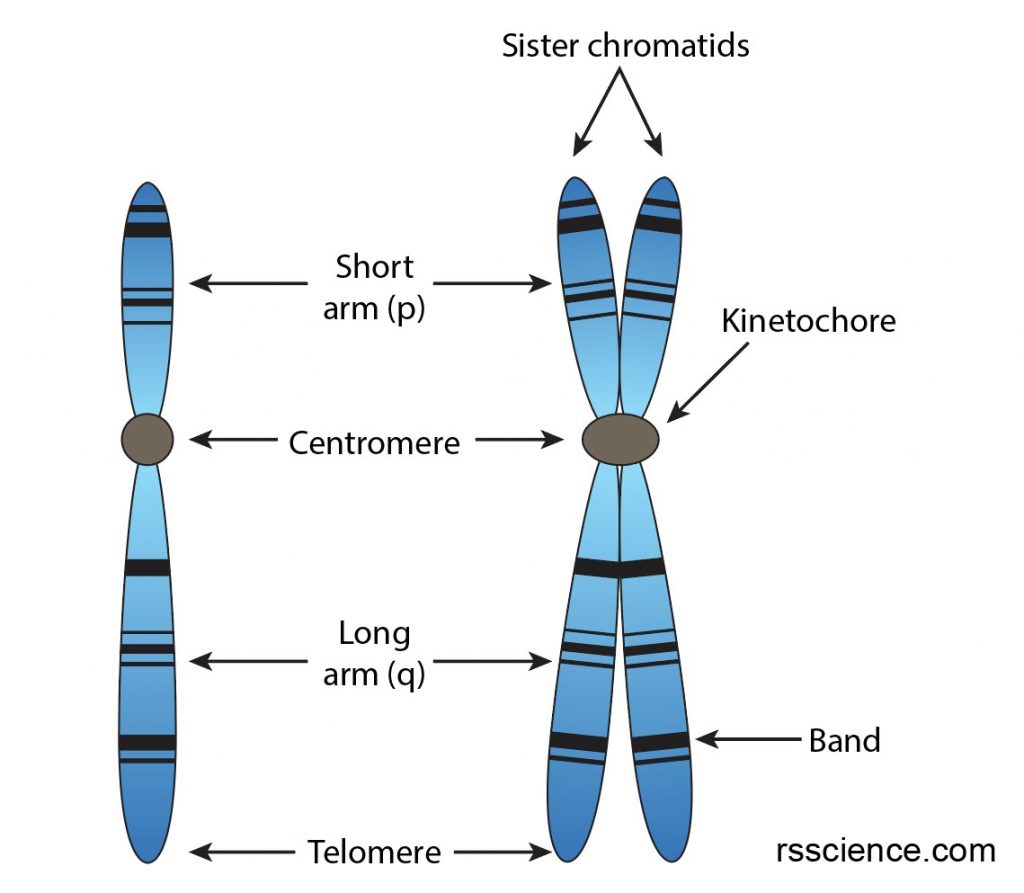
[In this image] The structure of chromosomes.
Before cell division, each chromosome replicates to form a sister chromatid, which has four arms attached to one centromere (also referred to as kinetochore).
Centromere
Although this constriction point is called the “centro – mere”, it is usually not located precisely in the center of the chromosome. In some cases, the centromere locates almost at the chromosome’s end. The location of the centromere on each chromosome gives the chromosome its characteristic shapes and can be used to describe the location of specific genes.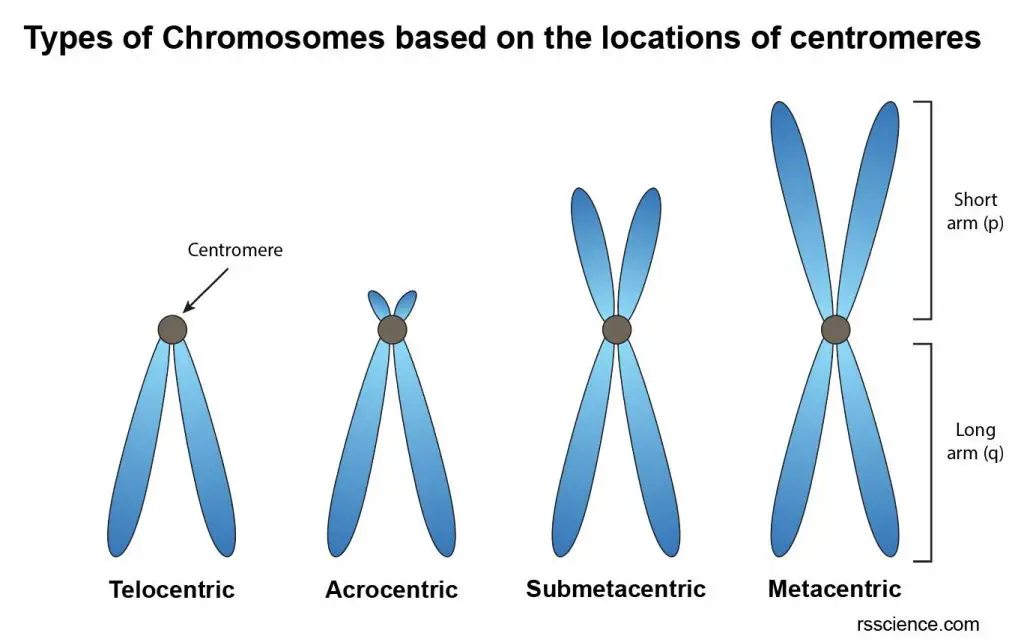
[In this image] Four types of chromosomes based on the location of centromeres.
(1) Telocentric; (2) Acrocentric; (3) Submetacentric; (4) Metacentric.
Based on the locations of centromeres, the chromosomes can be classified into four types:
(1) Telocentric – Its centromere occupies the terminal position so that the chromosome has just one arm. This type of chromosome doesn’t exist in human cells.
(2) Acrocentric – Its centromere locates closely to one of the ends. As a result, one arm is very long, and the other is very short.
(3) Submetacentric – Its centromere locates slightly away from the mid-point so that the two arms are unequal.
(4) Metacentric – Its centromere lies in the middle of chromosome so that the two arms are almost equal.
During cell division, the centromeres serve as the anchor points to pull sister chromatids apart and equally into two daughter cells. We will discuss more later in the section of “Chromosomes and Cell Division”.
A normal chromosome has only one centromere (called Monocentric). If a chromosome has two (Dicentric) or more than two centromeres (Polycentric), the separation of sister chromatids will be a problem. If a chromosome has no centromere, it’s called Acentric. Such chromosomes represent freshly broken segments of chromosomes that do not survive for long. These abnormal chromosomes are often the consequence of DNA recombination.
Karyotype
Scientists learned most of what scientists know about chromosomes by observing chromosomes during cell division. A special microscopic technique, called Giemsa banding (G-banding) karyotype, can show individual chromosomes like the image below.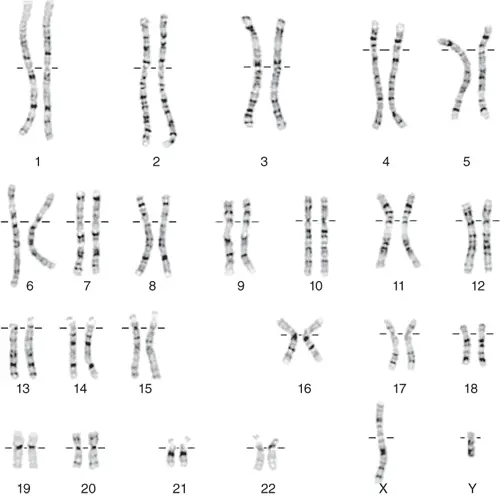
[In this image] A karyotype is an individual’s collection of chromosomes. As you can see here, this is a karyotype of a male (22 pairs of autosomes plus X and Y chromosomes).
These darker “G-bands” that you see on the chromosomes come from the stronger staining of Giemsa stain with more condensed chromatin. These regions tend to be rich with adenine and thymine (AT-rich) and relatively gene-poor. On the other hand, the lighter G-bands reflect less condensed chromatin with higher guanine and cytosine contents (GC-rich). These regions also carry more genes and are transcriptionally active.
Chromosome Banding and Nomenclature
Each chromosome arm is divided into regions or cytogenetic bands based on the G-band karyotype. The cytogenetic bands are labeled p1, p2, p3, or q1, q2, q3, etc., counting from the centromere out toward the telomeres. At higher resolutions, sub-bands can be seen within the bands. The sub-bands are also numbered from the centromere out toward the telomere.
Geneticists use these bands as maps to describe the location of a particular gene on a chromosome. This method is called Cytogenetic location. The combination of numbers and letters provides a gene’s “address” on a chromosome.
For example:
7q12 : A location on chromosome #17, region #1, band #2 of q arm.
7q31.2 : A location on chromosome #7, region #3, band #1 of q arm, sub-band #2
17q13-q22 : A segment on chromosome #17, from q13 to q22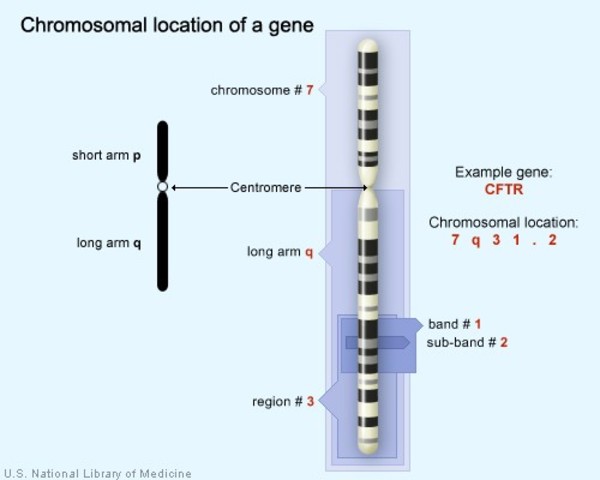
[In this image] The CFTR gene is located on the long (q) arm of chromosome 7 at position 7q31.2. The mutation of CFTR gene is responsible for a genetic disease called cystic fibrosis.
Image source: MedlinePlus
Sometimes, we label the ends of the chromosomes as “ptel” and “qtel”. For example, the notation 7qtel refers to the end of the long arm of chromosome 7. The gene very close to the centromere may sometimes be labeled with ” cen ” abbreviations.
Telomeres
A telomere is the end of a chromosome. Telomeres are made of repetitive sequences of non-coding DNA that protect the chromosome from damage. Each time a cell divides, the telomeres become shorter. Eventually, the telomeres become so short that the cell can no longer divide.
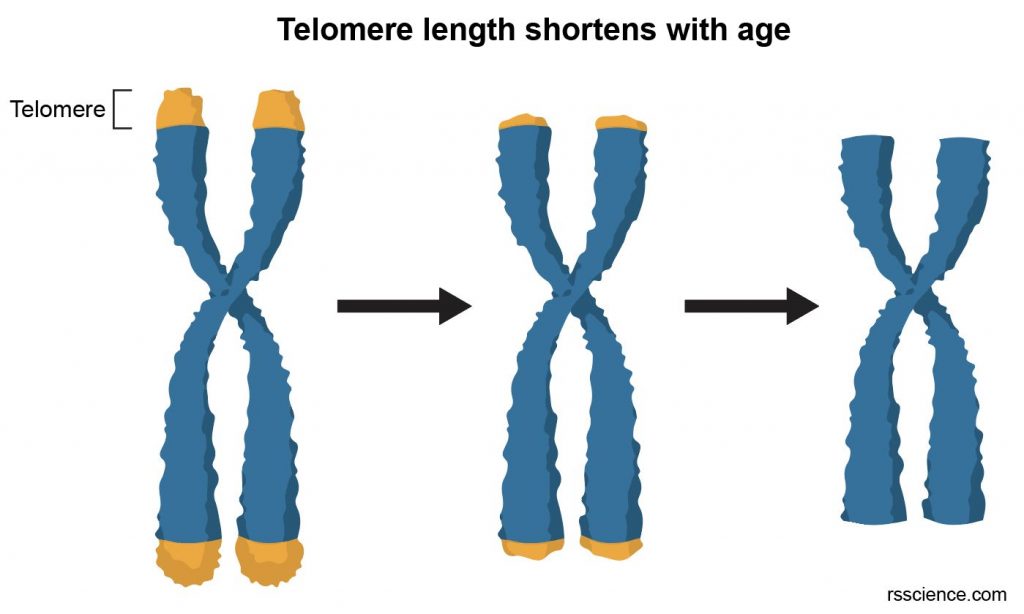
[In this image] Telomere length as a marker of biological age. Over time, due to ongoing cell division, telomeres become shorter. Cells will sense the lack of telomere length and stop division. Otherwise, chromosomes may start losing genes from the ends and create mutation.
Autosomes and sex chromosomes
In humans, each cell normally contains 23 pairs of chromosomes, for a total of 46. Twenty-two of these pairs, called autosomes, look the same in both males and females. The 23rd pair, the sex chromosomes, differ between males and females. Females have two copies of the X chromosome, while males have one X and one Y chromosome. Each parent contributes one chromosome to each pair so that offspring get half of their chromosomes from their mother and a half from their father.
Chromosomes and Cell Division
There are two types of cell division: mitosis and meiosis. Both require a proper separation of equal amounts of genetic materials or chromosomes into new daughter nuclei or cells.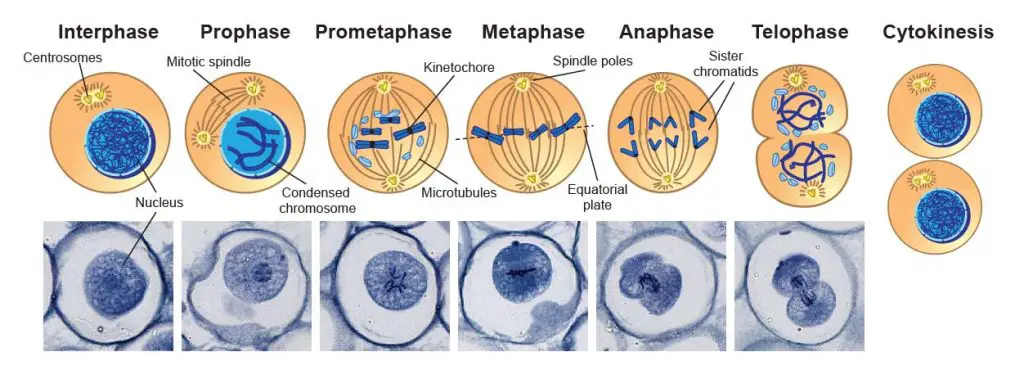
[In this image] Mitosis phases under the microscope.
Mitosis consists of five basic phases: prophase, prometaphase, metaphase, anaphase, and telophase. These phases occur in strictly sequential order. This specimen is Ascaris eggs (parasitic nematode worm) that contain active mitotic cells and can be purchased from Rs’ Science 25 Premade slide set.
Centromeres help to keep chromosomes properly aligned during the complex process of cell division. As chromosomes are copied in preparation for the production of a new cell, the centromere (or sometimes, called kinetochore) serves as an attachment site for the two halves of each replicated chromosome, known as sister chromatids.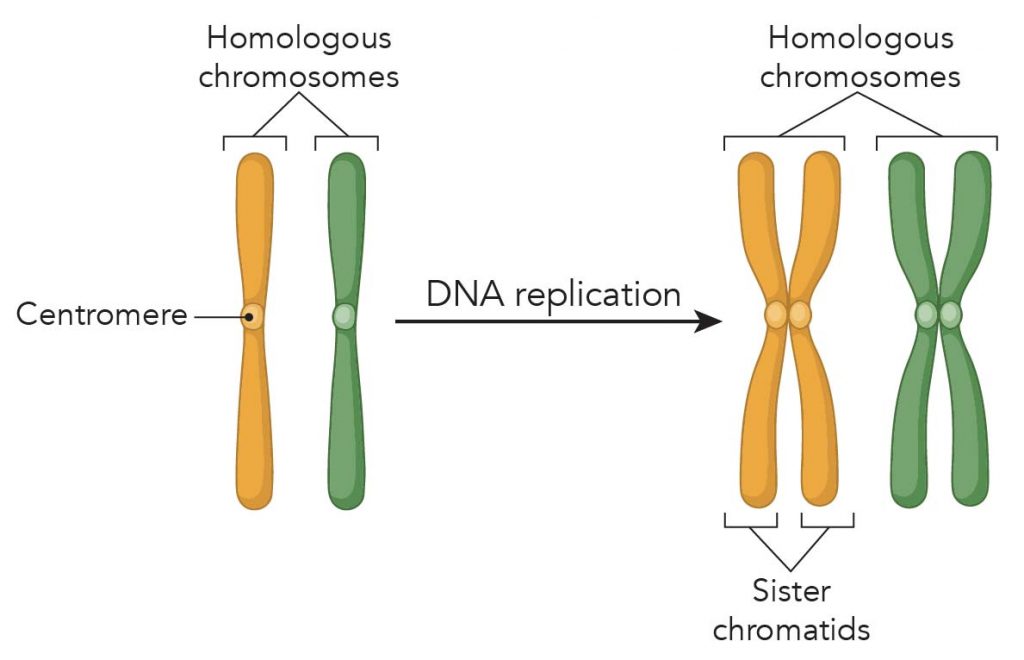
[In this image] Chromosome replication forms sister chromatids. The X-shape sister chromatid has four arms attached at a centromere.
Later in the phases of mitosis, the sister chromatids have to be pulled apart and separated equally into two new nuclei. Cells do this job exactly like how we pull a car out of mud using a cable and a winch. The “cable” is called “microtubule”. The centromeres serve as an anchor site for microtubules. The centrosome works like a “winch” pulling the sister chromatids (the car) apart.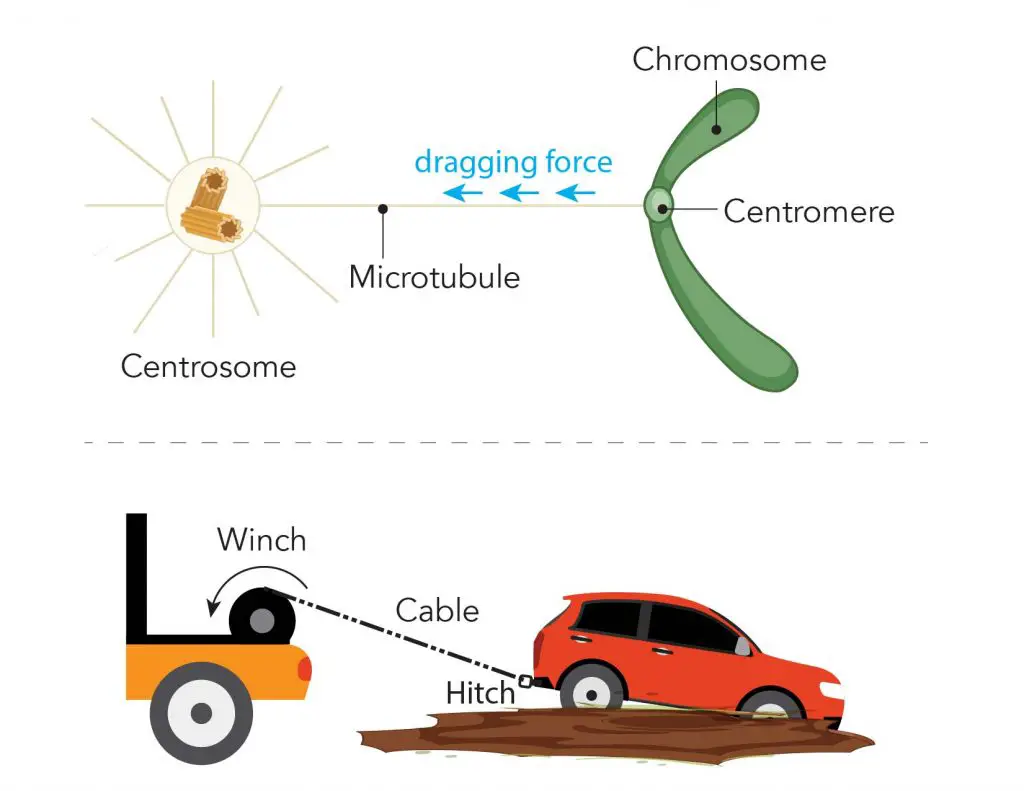
[In this image] Illustration of microtubule, centromere, and centrosome during mitosis.
During mitosis, one end of the microtubule is attached to the centromere regions of each chromosome, and the other ends are controlled by centrosomes. You can imagine that the centrosome like the winch pulls the sister chromatids (the car) apart by dragging the microtubule (the cable) anchored to the centromere (the tow hitch).
After cell division, the chromosomes will disassemble into more loose DNA threads in the new daughter cells. Opening up the packed chromatin allows the gene transcription for the daily activities of cells.
How to pack DNA into chromosomes
To assemble a chromosome, cells make special proteins called histones that can condense DNA. The resulting histone-DNA complex is called chromatins.
If you ever tried to store sewing thread, you know that it is much easier to coil the thread on a cylindrical roll. You imagine that DNAs are like sewing threads and the histones are like cylindrical rolls. You got my idea.
In brief, eight histone proteins form a core complex, called nucleosome. Each nucleosome allows two turns of DNA wrapped. Under an electron microscope, the DNA with nucleosomes looks like many beads on a string.
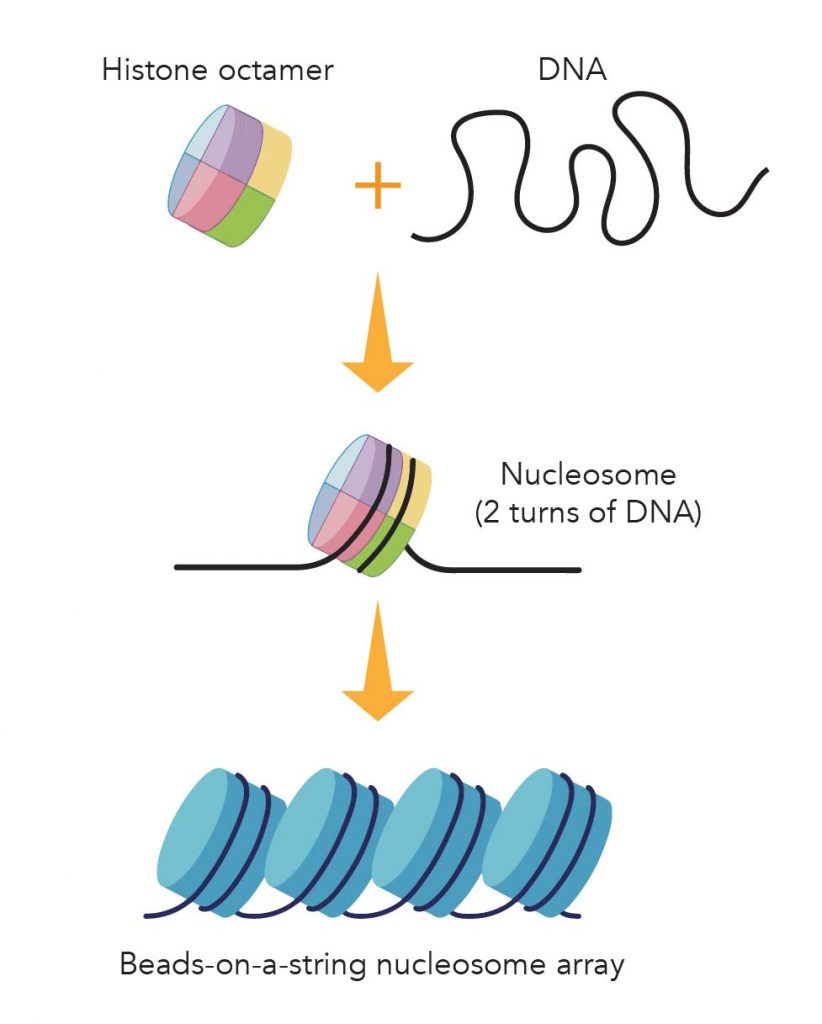
[In this image] Illustration of the package of DNA threads by histone.
Eight core histone proteins form a histone octamer. About 147 base-pairs of DNA wraps around the histone octamer (~ two turns) to form a nucleosome. Nucleosomes arrayed along the DNA molecules look like beads on a string.
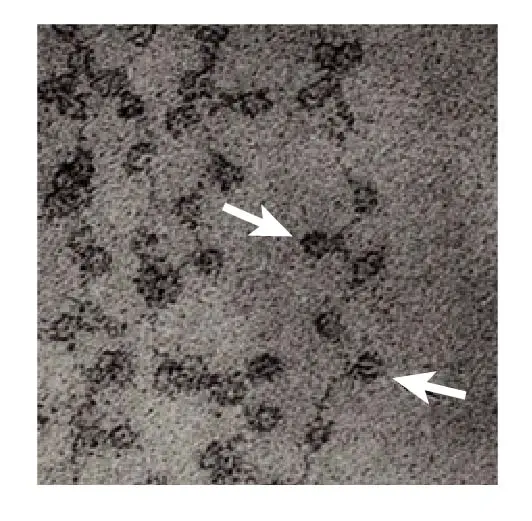
[In this image] Electron microscopic image of DNA wrapping around the nucleosomes like the beads on a string.
The nucleosomes are indicated by arrows.
Image modified from Olins, D. E. & Olins, A. L. Chromatin history: our view from the bridge. Nature Reviews Molecular Cell Biology 4, 811 (2003).
Packaging DNA into nucleosomes shortens the DNA thread length about sevenfold. These nucleosomes then are further packaged into the most compact rod-shaped structures of chromosomes.
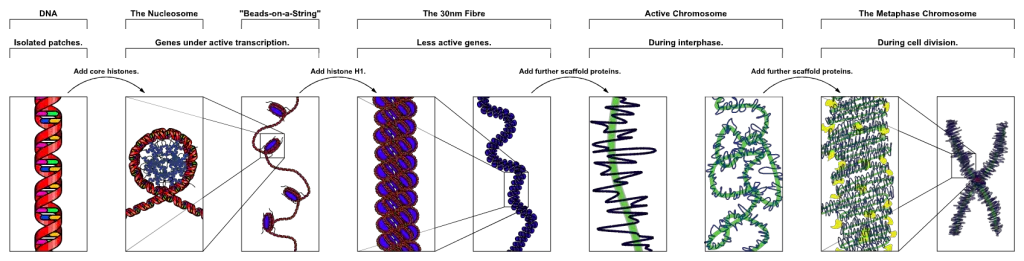
[In this image] The process of chromosome package.
The DNA wrapped around histones and together with scaffold protein to form a chromosome. Currently, scientists only have a good idea about how DNA organizes into nucleosomes. Beyond that, we still don’t know exactly how chromosomes assemble.
Image credit: wiki
Chromosome aberrations and diseases
Chromosomal aberrations, or abnormalities, are changes in the number or structure of chromosomes. Chromosomal aberrations are most often caused by errors during cell division. Since chromosomes are the carriers of our genes, a chromosomal aberration can cause severe medical disorders.
Wrong chromosomal numbers
Having missed or extra chromosomes is called aneuploidy. The condition with an extra chromosome (three instead of two) is trisomy. For example, Down syndrome is a disorder due to the presence of an extra chromosome 21, or “trisomy 21”. Aneuploidies of the sex chromosomes can also occur like XXY, XYY, and XXX syndromes.
Wrong chromosomal structure
Errors during cell division may change the structures of chromosomes and mess up our genes. There are four types of structural chromosomal aberrations: deletion, duplication, inversion, and translocation.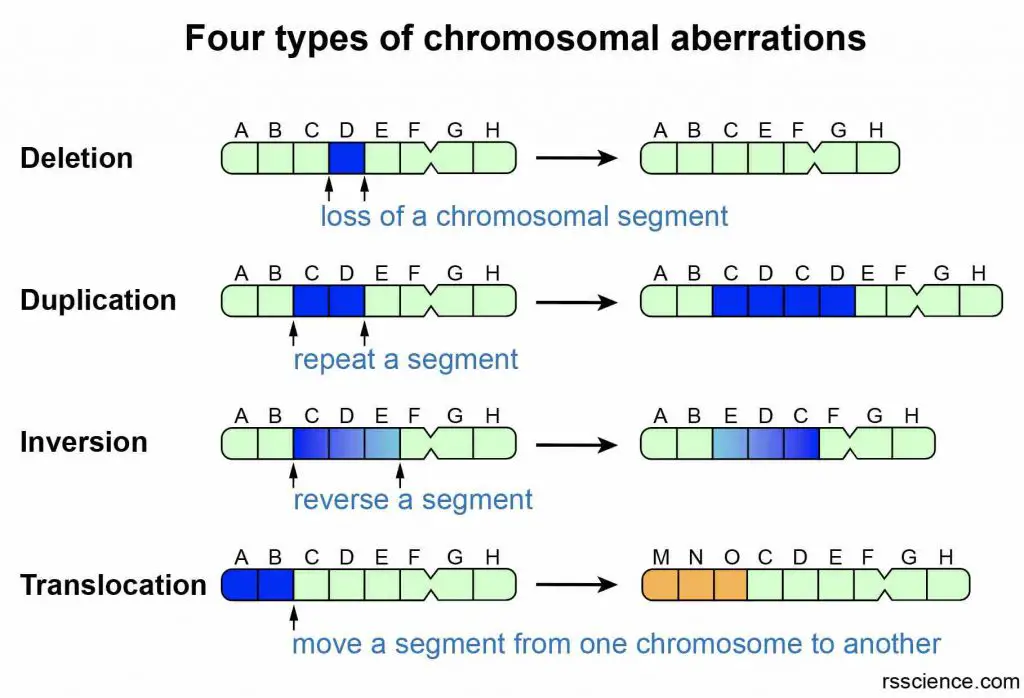
[In this figure] Four types of structural chromosomal aberrations.
(1) Deletion: Loss of genes from a chromosome. (2) Duplication: Genes are doubled in number on a chromosome. (3) Inversion: A segment of a chromosome is broken and gets reattached to the same chromosome in an inverted (180° twist) position. There is no loss or gain of genes. (4) Translocation: Transposition of a part of a chromosome to a non-homologous chromosome.
The discovery of Chromosomes
As early as the 1860s, Gregor Mendel and Charles Darwin began to explore possible mechanisms of heredity. However, they didn’t know the existence of genetic materials.
Then, over the next few decades, Walther Flemming, Theodor Boveri, and Walter Sutton made a series of important discoveries around chromosomes, including what these structures looked like, how they moved during mitosis, and what role they likely played in heredity.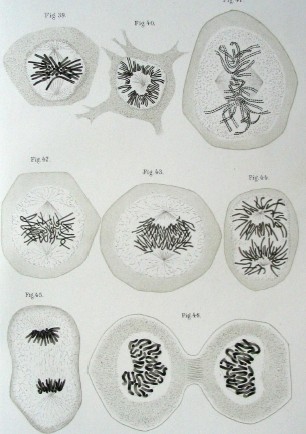
[In this image] Drawing of mitosis by Walther Flemming from the book Zellsubstanz, Kern und Zelltheilung, 1882
Image source: wiki
The name “chromosome” came from the Greek “chroma” (meaning color) and “-soma” (meaning body), describing that the chromosomes can be stained with dyes in the nuclei.
Flemming had actually discovered the chromosome, although the term would be coined a few years later by Heinrich Waldeyer.
Chromosome numbers in different organisms
The number of chromosomes in eukaryotes is highly variable (see examples below). There is no rule that a more advanced organism has more chromosomes (or genes) than a lower organism. In fact, chromosomes can fuse or break and thus evolve into different numbers in closely related species.
Chromosomes in animal cells are usually in pairs (called diploids or 2n). However, in plants, chromosomes could be in the copies of 4 (tetraploid), 6 (hexaploid), or more (polyploid). The Indian muntjac, a deer species native to South Asia, has the lowest recorded number of chromosomes (6 for females and 7 for males) of any mammal. The Adder’s-tongue ferns may have the largest number of chromosomes (approx. 1,200; polyploid) in the known plant kingdom.
Some birds, reptiles, fish, and amphibians (not mammals) have a type of very small chromosome, called microchromosome. For example, chicken has 8 large chromosomes, 2 sex chromosomes, and 60 microchromosomes.
[In this image] A few examples showing the diversity of chromosome numbers in animals and plants.
Do Prokaryotes have chromosomes?
Yes, prokaryotes – bacteria and archaea – typically have a single circular chromosome. Since prokaryotes do not have nuclei, and instead, their DNA is organized into a structure called the nucleoid. They have histone-like proteins to help the organization of DNA. Sometimes, bacteria also have smaller circular extrachromosomal DNA called plasmids.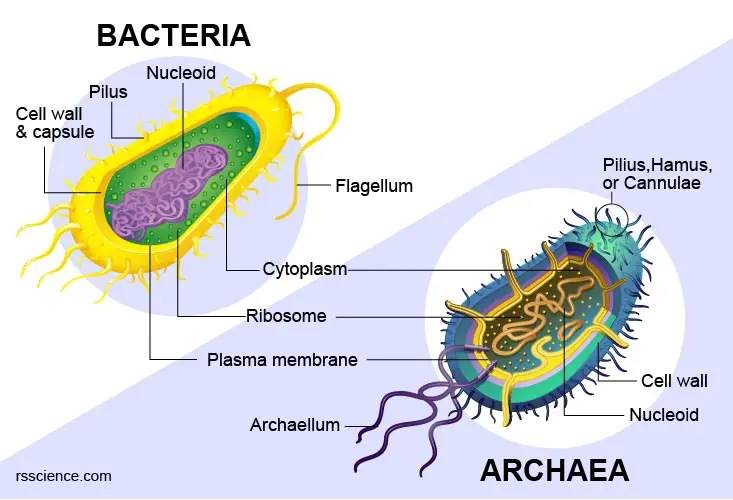
[In this image] Structural similarities and differences between bacterial and archaeal cells.
Learn more: https://rsscience.com/archaea-vs-bacteria/
Summary
1. Chromosomes are made up of bundles of tightly coiled DNA molecules and proteins called histones. In eukaryotic cells, chromosomes are stored in the nuclei.
2. DNA threads are too thin (2.2-2.6 nm) to be seen with a light microscope. However, the chromosomes can be seen with a light microscope during cell division.
3. We have 23 pairs of chromosomes (1-22, X, and Y), and the number will double right before the cell division.
4. Each chromosome has a constriction point called the centromere, which divides the chromosome into two arms. The short arm of the chromosome is labeled the “p arm.” The long arm is labeled the “q arm.” The ends of the chromosome are called telomeres.
5. Chromosomes in animal cells are usually in pairs (called diploids or 2n). However, in plants, chromosomes could be in the copies of 4 (tetraploid), 6 (hexaploid), or more (polyploid).
6. Chromosome number is variable from eukaryotes to eukaryotes. More chromosomes in number doesn’t correlate to the more advanced organism.