If DNA is a book, then how is it read? It requires two skills – transcription and translation. In this article, you will learn more about the transcription process, where DNA is converted to RNA, a more portable set of instructions for the cells.
This article covers
Overview of transcription
Transcription is the process of making an RNA copy of a DNA segment. This copy is called a messenger RNA (mRNA) molecule. This mRNA will be transported from the cell nucleus to the ribosomes in the cytoplasm, where the mRNA directs the synthesis of the protein.
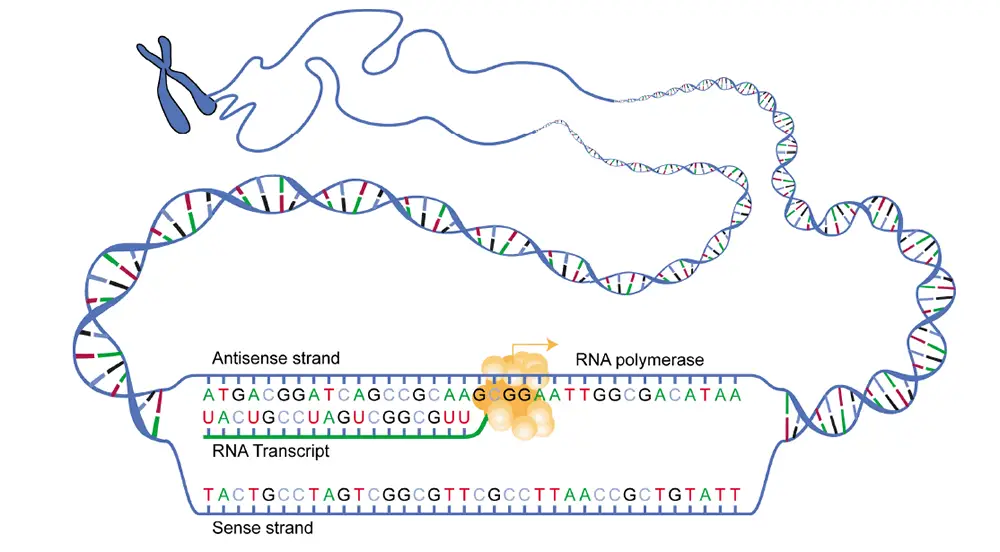
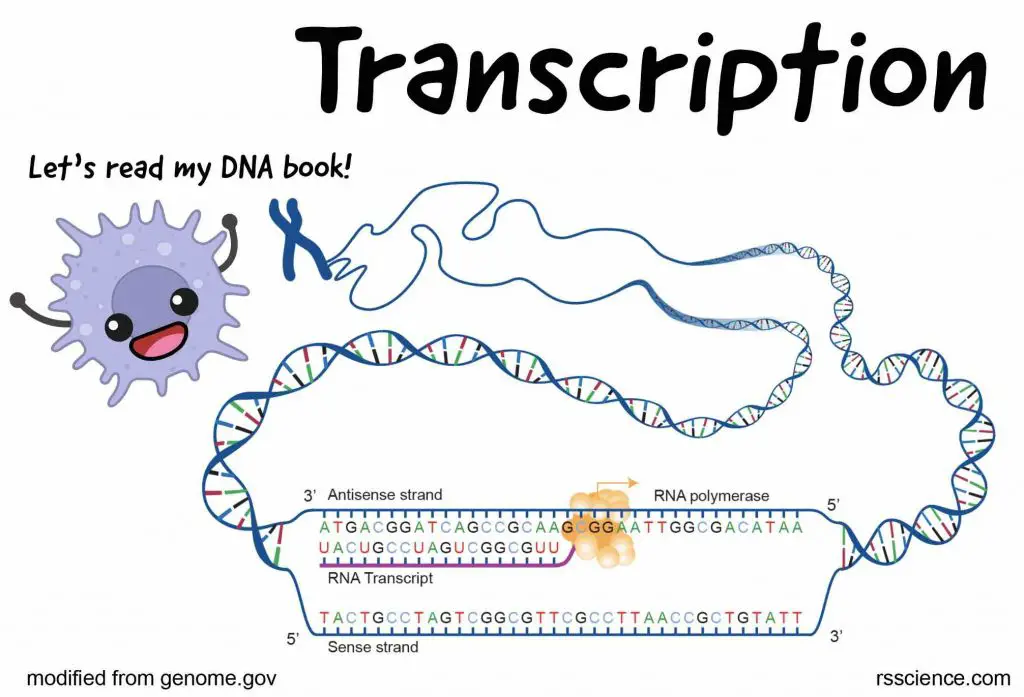
[In this image] Overview of transcription.
You can see in this diagram that a piece of unwound DNA with an RNA polymerase working on the transcription: making an RNA transcript from a DNA template.
Image source: NIH
Gene expression
Genes contain the information to build proteins that cells need. Our genes are written as the nucleotide base pairs (A, T, G, C) in the DNA. For a gene to exert its function, the genetic information must read out to build a protein. This process is called gene expression.
There are two steps for making proteins from genes:
First, inside the cell nucleus, the transcription makes copies of a particular gene in the form of massager RNAs (mRNAs). The RNA copy of a gene’s DNA sequence is also called a transcript, which carries the information needed to build a protein.
Second, these mRNAs are exported outside the nucleus to the cytoplasm for ribosomes to make proteins. Since RNA and proteins’ languages are very different, the ribosomes have to decode the genetic information. Therefore, we called this step translation.
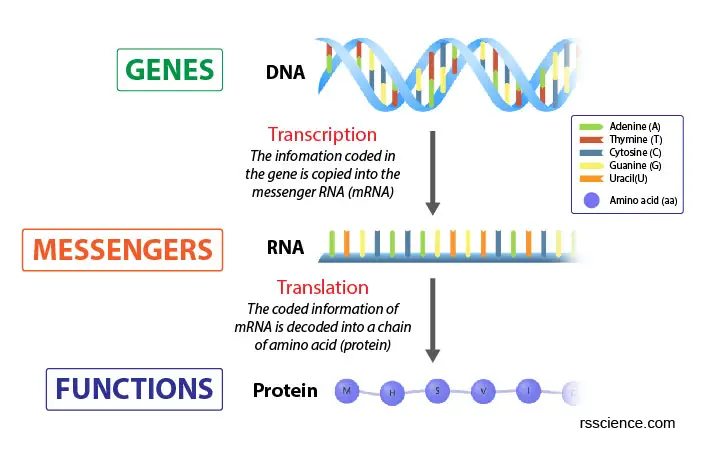
[In this image] The Central Dogma of Biology explains the flow of genetic information within a biological system.
It is often stated as “DNA makes RNA, and RNA makes protein”, or in short, “DNA → RNA → Protein”. Transcription is responsible for the first part of going from DNA to RNA. Translation is the second process of making protein from RNA.
What does “Transcription” mean?
Transcription is a process in which information is rewritten. For example, you took draft notes in class, and then you rewrote them neatly in a notebook to help you review. Or your friend left a message on your voicemail, and you had to rewrite it down on paper. If you made a mistake while you rewrote or “transcribed” the critical information (i.e., phone number, address, next exam date … etc.), it would be bad.
In biology, transcription is the process of rewriting the information of genes from the DNA sequence to the form of RNA. You can imagine that our genome is a huge library storing all our genetic information safely inside the nucleus. One day, our cell wants to make more hemoglobin proteins; it goes to the library, finds the hemoglobin blueprint book, and makes a copy. Now, our cell can mail this portable copy, instead of the whole library, to the ribosomes in the cytoplasm. Finally, the ribosomes read the instruction and manufacture the requested hemoglobin proteins.
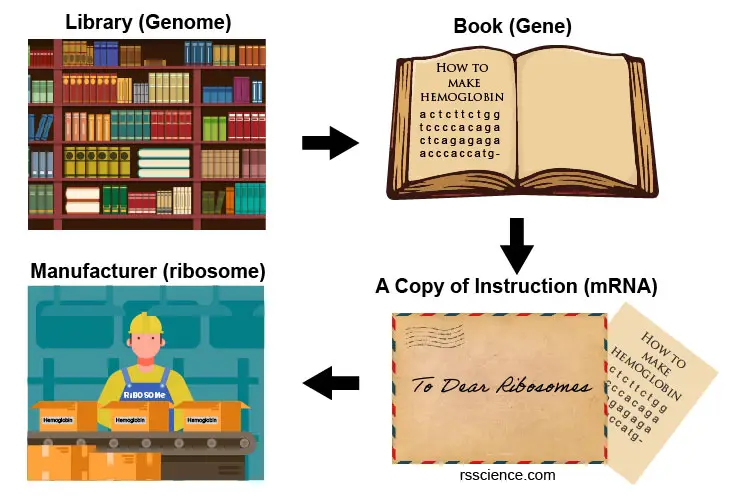
[In this image] How to place an order of hemoglobin?
(1) Going to the whole genome library in the nucleus. (2) Find the book named “The Ultimate Guide of Making Hemoglobin”. (3) Find the pages of the hemoglobin coding sequence. (4) Make a copy of the instruction. Remember to change the letters “T” to “U”. (5) Mail the copy to the ribosomes in the cytoplasm. (6) Ribosomes read the codes and produce hemoglobins accordingly.
To sustain our everyday lives, our cells must repeat this process (gene expression) 24 hours a day, 7 days a week. Transcription is vital to our cells and our body’s overall health; therefore, it must be done 100% error-free! Let’s see how our cells transcribe thousands of million DNA-RNA alphabets per day without making a mistake.
The language of DNA – A, T, G, C
Both DNA and RNA are long-chain molecules of nucleotides. Each nucleotide is made up of three components: a nitrogen-containing base, a five-carbon sugar, and a phosphate group. The nitrogenous base is either a purine or a pyrimidine. The five-carbon sugar is either a ribose (in RNA) or a deoxyribose (in DNA) molecule.
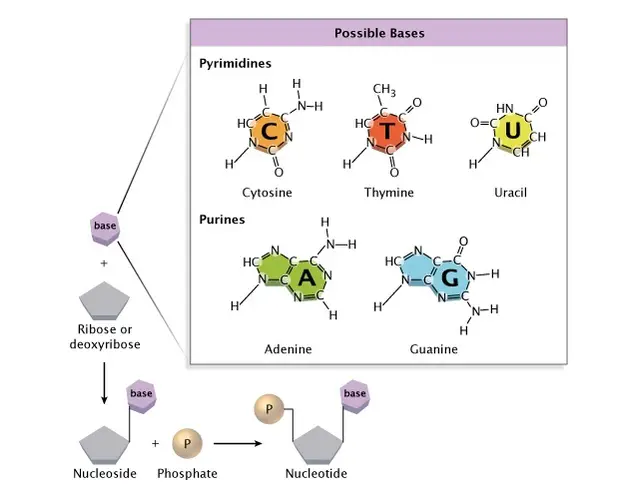
[In this image] The chemical structure of a nucleotide.
Image source: nature
The DNA alphabets contain only four letters, which are four nitrogenous base options: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). Each base can only bond with one other, A with T and C with G. This is called the DNA complementary base pairing rule. By this rule, two complementary DNA strands pair together and form a double-stranded DNA helix.
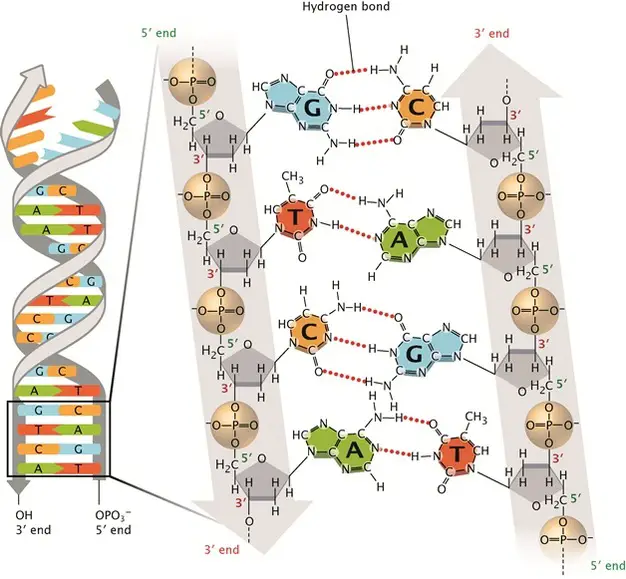
[In this image] DNA complementary base pairing results in the DNA double-stranded helix.
Two hydrogen bonds connect T to A; three hydrogen bonds connect G to C. The sugar-phosphate backbones (grey) run anti-parallel to each other so that the two DNA strands are aligned.
Image source: nature
Directionality of DNA strands
As shown in the diagram above, the two ends of a strand of DNA or RNA strand are different. That is, the directionality to read a DNA or RNA strand.
At the 5’ end of the chain, the phosphate group of the first nucleotide in the chain sticks out. The phosphate group is attached to the 5′ carbon of the sugar ring, which is why this is called the 5′ end.
At the 3’ end, the hydroxyl of the last nucleotide added to the chain is exposed at the other end. The hydroxyl group is attached to the 3′ carbon of the sugar ring, which is why this is called the 3′ end.
Many processes, such as DNA replication and transcription, can only take place in one particular direction, from 5’ end to 3’ end.
The Language of RNA – A, U, G, C
To transcript DNA into mRNA, the rule is the same. The only difference is that Uracil (U) replaces Thymine (T). So, G ↔ C, A → U, and T → A. In our cell, the transcription is done by an enzyme called RNA polymerase in the nucleus, which can synthesize mRNA from a DNA template.
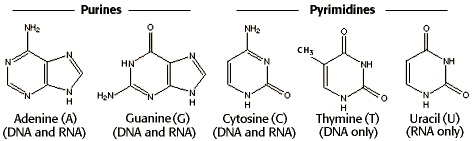
[In this image] Nitrogenous base options for DNA and RNA.
Image source: wiki
Practice the transcription of DNA to RNA
By knowing the DNA-RNA complementary base pairing rule, you can determine the complementary strand to a single DNA strand based only on the base pair sequence. For example, let’s say you know the sequence of one DNA strand that is as follows:
DNA (coding strand): 5’-TTG ACG ACA AGC TGT TTC-3’
Using the complementary base pairing rules, you can conclude that the complementary strand is:
DNA (template strand): 3’-AAC TGC TGT TCG ACA AAG-5’
RNA strands are also complimentary except that RNA uses U instead of T. Therefore, you can also infer the mRNA strand that would be produced from the first DNA strand. It would be:
mRNA: 5’-UUG ACG ACA AGC UGU UUC-3’
This mRNA molecule will then be translated to a protein in the ribosome by a much complicated “genetic codon” rule. To learn this part, visit our article about “How to Read the Amino Acids Codon Chart? – Genetic Code and mRNA Translation.”
What kinds of RNA can be transcribed (produce)?
When talking about RNA and transcription, we usually refer to messenger RNA (mRNA). An mRNA is transcribed from a protein-encoding gene and subsequently is translated to a protein. But there is a whole set of other RNAs that get transcribed, like transfer RNA (tRNA) and ribosomal RNA (rRNA), that do different functions in the cells.
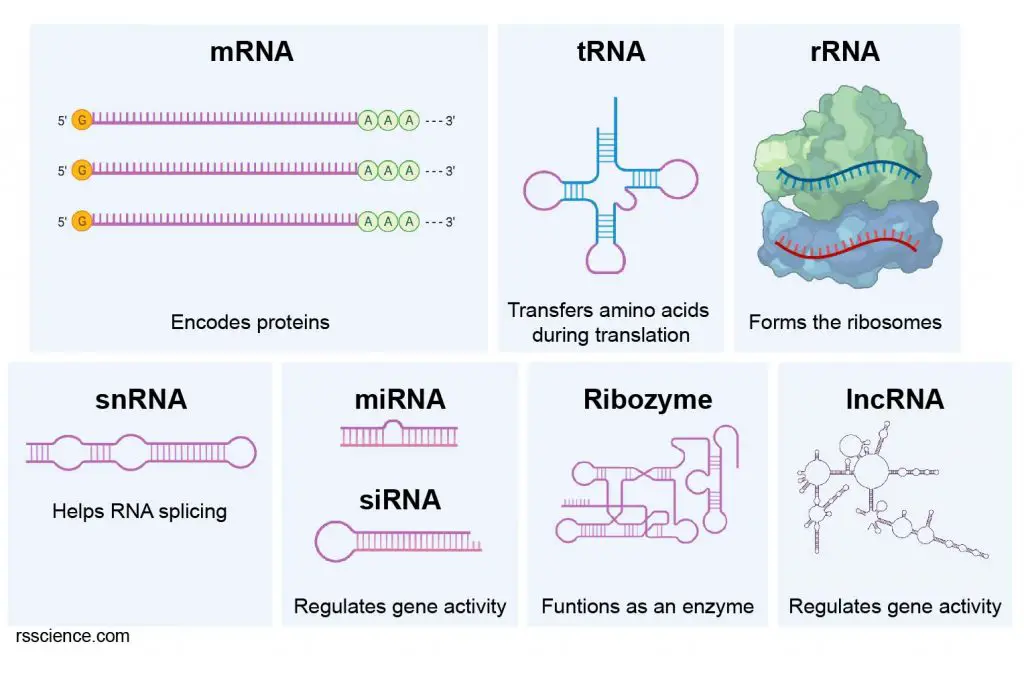
[In this image] Types of RNA molecules.
Transcription produces RNAs that functionally are either for protein-coding (mRNA) or non-coding (so-called “RNA genes“). At least six functional types of RNA genes exist:
1. Transfer RNA (tRNA) — During translation, tRNAs transfer specific amino acids to the growing polypeptide chains in the ribosomes for protein synthesis.
2. Ribosomal RNA (rRNA) — rRNAs and ribosomal proteins assemble into ribosomes.
3. Small nuclear RNA (snRNA) — snRNAs help the splicing of pre-messenger RNAs in the nucleus.
4. Micro RNA (miRNA) — miRNAs can regulate gene activity. Some miRNAs bind to mRNAs and block the translation. Scientists use the same principle to synthesize small interfering RNA (siRNA), which can be used as a drug.
5. Catalytic RNA (ribozyme) – Ribozymes are RNA molecules but have enzymatical activities.
6. Long non-coding RNAs (lncRNA) – lncRNAs are RNA transcripts of more than 200 nucleotides that are not translated into any protein. lncRNA was discovered to regulate gene functions. However, the actual mechanisms are still unclear.
Note: Different types of RNAs are transcribed by different RNA polymerases. We will cover this topic in the next section.
RNA polymerase
RNA polymerase is an enzyme that is responsible for copying a DNA sequence into an RNA sequence, during the process of transcription.
RNA polymerase uses single-stranded DNA as a template to synthesize a complementary RNA strand. RNA polymerase builds the RNA strand by adding new nucleotide units one by one. For instance, if there is a “G” in the DNA template, RNA polymerase will add a “C” to the new, growing RNA strand. In addition, the new RNA strand extends in the direction of 5′ to 3′. RNA polymerase adds new nucleotides to the 3′ end of the RNA strand.
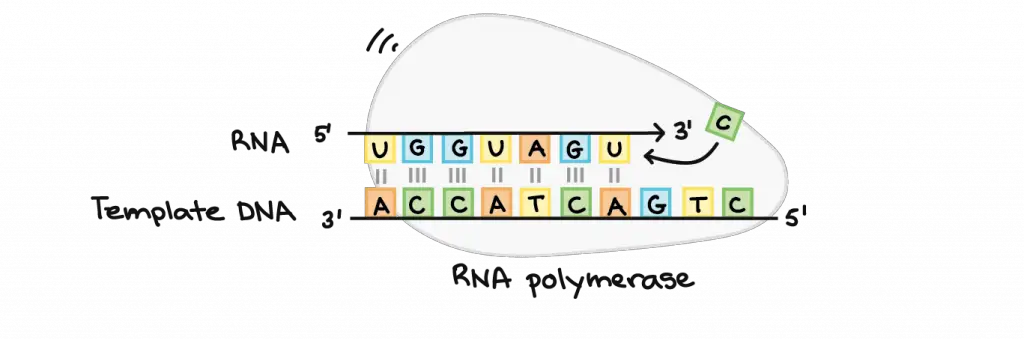
[In this image] The action of RNA polymerase.
According to the “G” in the DNA template, the RNA polymerase adds a “C” to the 3′ end of the RNA strand.
Image credit: Khan Academy

[In this image] The 2006 Nobel Prize in Chemistry was awarded to Roger D. Kornberg for understanding the action of RNA polymerase.
Photo credit: The Nobel Prize
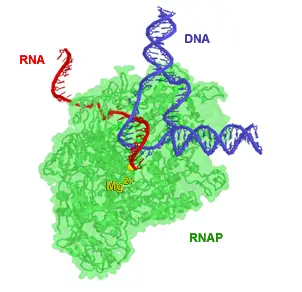
[In this image] Structural model of RNA polymerase in action: RNA polymerase (green) unwinds the DNA double helix (blue) and uses one strand as a template to create the single-stranded messenger RNA (red). The magnesium ion (yellow) is located at the enzyme active site.
Image source: wiki
RNA polymerase is essential to life and is found in all living organisms and many viruses. However, the number and composition of RNA polymerase vary across domains. For instance, bacteria contain a single type of RNA polymerase, while eukaryotes have three distinct types:
1. RNA polymerase I synthesizes ribosomal RNA (but not 5S rRNA, which is synthesized by RNA polymerase III).
2. RNA polymerase II synthesizes precursors of mRNAs and most small RNA and micro RNAs.
3. RNA polymerase III synthesizes tRNAs, rRNA 5S, and other small RNAs found in the nucleus and cytosol.
Chloroplasts and mitochondria have their own unique RNA polymerases as well. However, chloroplast and mitochondrial RNA polymerases consist of a single protein unit. Conversely, all other RNA polymerases are protein complexes containing multiplex subunits.
Despite these differences, there are striking similarities among transcription processes in all lives, sharing a similar core structure and mechanism.
Stages of transcription
Transcription of a gene takes place in three stages: initiation, elongation, and termination. Here, we will briefly see how these steps happen.
Initiation
Near the beginning of each gene, there is a region of DNA sequence called the promoter. RNA polymerase has to find and bind to the promoter to initiate transcription. Many other proteins such as transcription factors and DNA sequences (like enhancers or suppressors) will also involve in deciding if an RNA polymerase can bind to a particular promoter or not. Once bound, RNA polymerase unwinds the DNA helix, providing the single-stranded template needed for transcription.
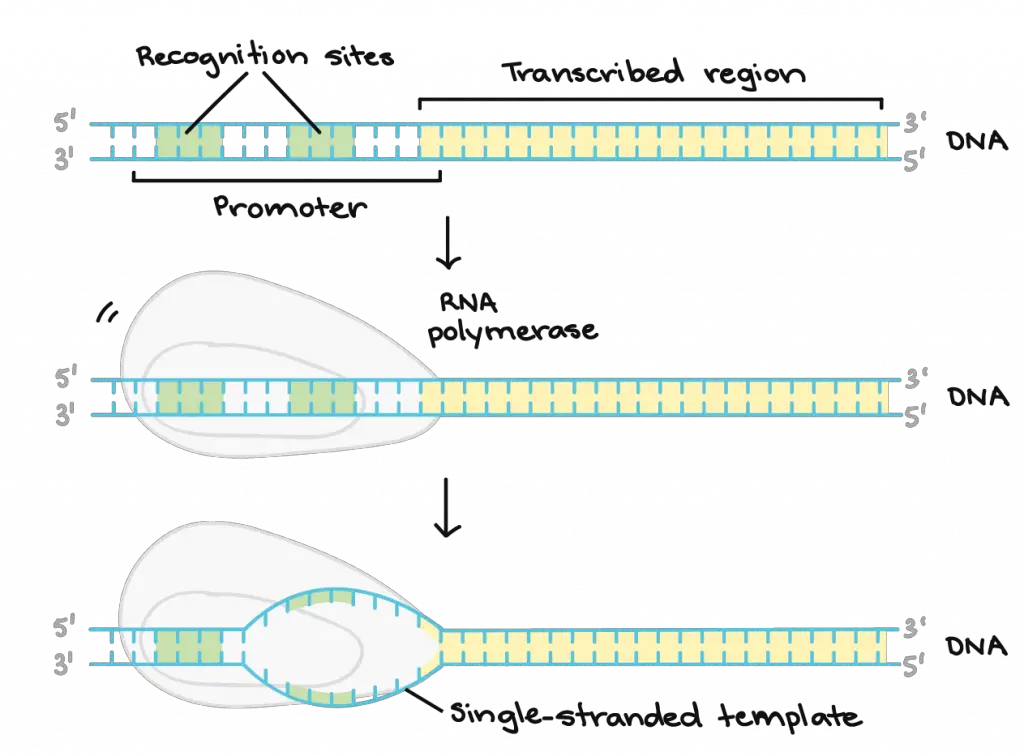
[In this image] Initiation of Transcription.
For most eukaryotic genes, their promoter regions come before (and slightly overlaps with) the transcribed region. The promoter region contains recognition sites for RNA polymerase or its helper proteins to bind to. The DNA opens up in the promoter region so that RNA polymerase can begin transcription.
Image credit: Khan Academy
Elongation
Once the DNA helix is opened, RNA polymerase starts “read” the DNA template strand one base at a time. It moves forward along the template strand in the 3′ to 5′ direction, opening the DNA double helix as it goes.
At the same time, RNA polymerase builds an RNA molecule out of complementary nucleotides, making a chain that grows from 5′ end to 3′ end. The synthesized RNA transcript only remains bound to the template strand for a short while, then exits the polymerase as a dangling string, allowing the DNA to close back up and form a double helix.
As a result, the RNA transcript copies the same information as the non-template (coding) strand of DNA, but it contains the base uracil (U) instead of thymine (T).
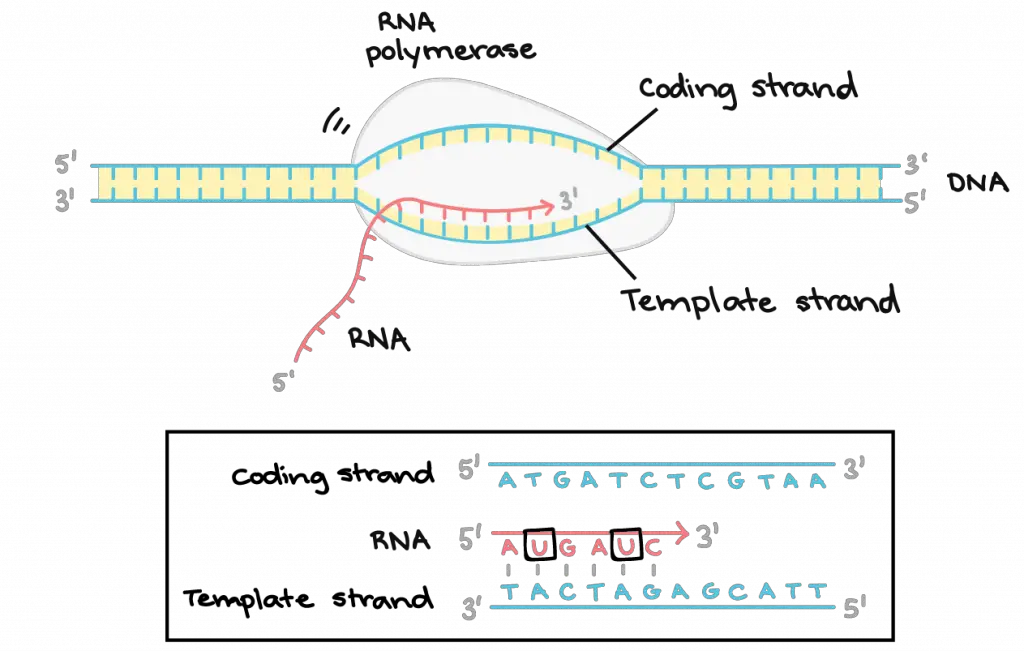
[In this image] Elongation of Transcription.
RNA polymerase synthesizes an RNA transcript complementary to the DNA template strand in the 5′ to 3′ direction. The “bubble” of opened DNA-RNA-RNA polymerase complex moves forward as the transcription proceeds.
Image credit: Khan Academy
Termination
RNA polymerase keeps reading and transcribing the DNA template strand until it reaches the sequences called terminators. The terminators signal that the RNA transcript is complete and cause the RNA transcript to be released from the RNA polymerase. RNA polymerase also closes up the DNA helix and detaches from the DNA molecules.
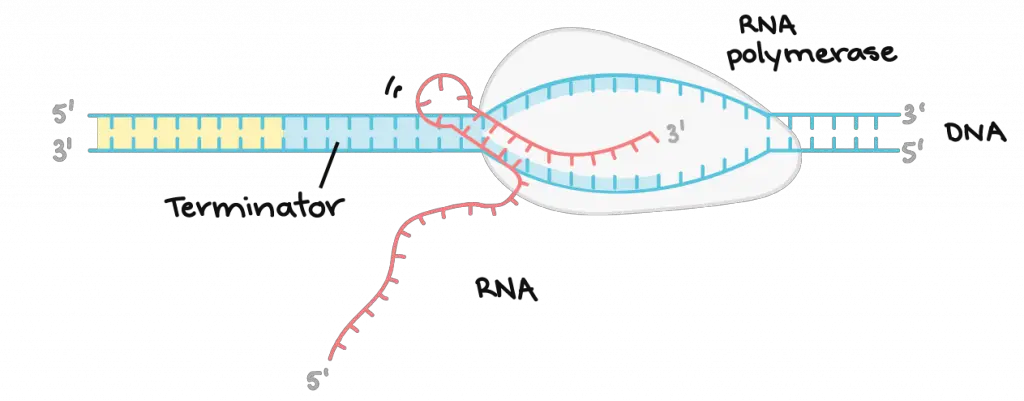
[In this image] Termination of Transcription.
The process of ending transcription is called termination, and it happens once the polymerase transcribes a sequence of DNA known as a terminator. Termination is slightly different in bacteria from in eukaryotic cells. This diagram shows an RNA hairpin as a termination signal for bacteria. In eukaryotes like humans, transcription termination happens when a polyadenylation signal appears in the RNA transcript.
Image credit: Khan Academy
Eukaryotic RNA modifications after transcription
In prokaryotic cells, the RNA transcripts can act as messenger RNAs (mRNAs) right away. However, in eukaryotic cells, the RNA transcripts produced by transcription are only pre-mRNAs. These pre-mRNAs must go through extra processing before they can be used in translation.
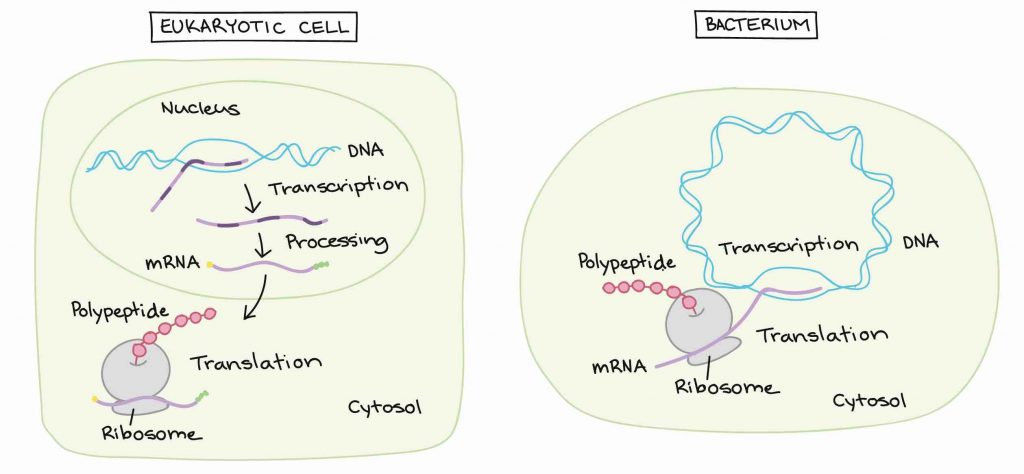
[In this image] The difference in gene expression between eukaryotic and prokaryotic cells.
In eukaryotic cells, mRNAs are transcribed and processed inside the nuclei. In prokaryotic cells, transcription and translation happen simultaneously in the cytosol.
Image credit: Khan Academy
Eukaryotic pre-mRNA processing includes:
A 5′ cap is added to the beginning of the RNA transcript, and a 3′ poly-A tail is added to the end.

[In this image] Diagram of a pre-mRNA with a 5′ cap and 3′ poly-A tail.
The 5′ cap is on the 5′ end of the pre-mRNA and is a modified G nucleotide. The poly-A tail is on the 3′ end of the pre-mRNA and consists of a long string of A nucleotides (only a few of which are shown).
Image credit: Khan Academy
5′ cap
The 5’ cap is added to the first nucleotide of the transcript during transcription. The cap is a modified guanine (G) nucleotide, and it protects the transcript from being broken down. It also helps the ribosome attach to the mRNA and start the translation.
Poly A tail
The poly-A (polyadenylation) tail is added to the 3’ end of the pre-RNA transcript immediately after transcription. The poly-A tail makes the RNA transcript more stable and helps it get exported from the nucleus to the cytosol. The number of adenine (A) nucleotides in each tail could be as many as 100-200.
Splicing
Many eukaryotic pre-mRNAs undergo splicing. In splicing, some sections of the RNA transcript (called introns) are chopped out, and the remaining sections (called exons) are pieced back together.
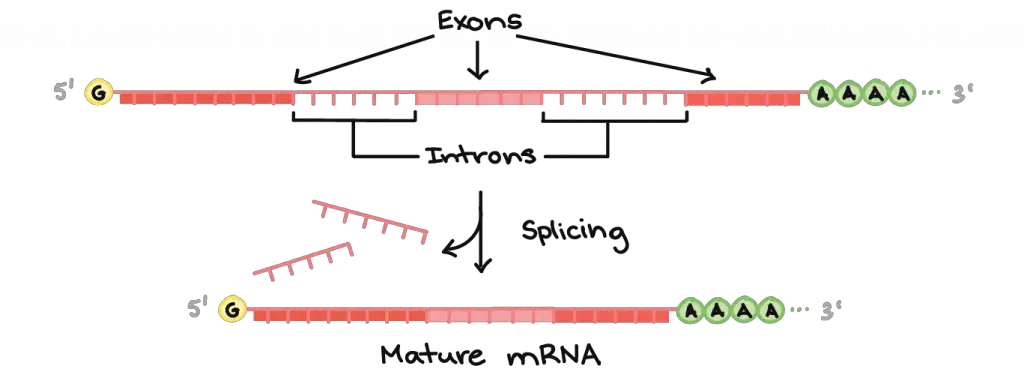
[In this image] Eukaryotic genes are not as compact as prokaryotic ones.
There are non-coding “introns” sitting in-between protein-coding segments or “exons”. The pre-mRNA still contains both exons and introns. The introns are removed from the pre-mRNA during splicing, and the exons are piece together to form a mature mRNA.
Image credit: Khan Academy
Some genes have multiple options of splicing (called alternative splicing). Like playing Lego bricks, alternative splicing produces different mature mRNA molecules (by assembling different exons) from the same initial transcript. As a result, one gene may produce several different versions of proteins.
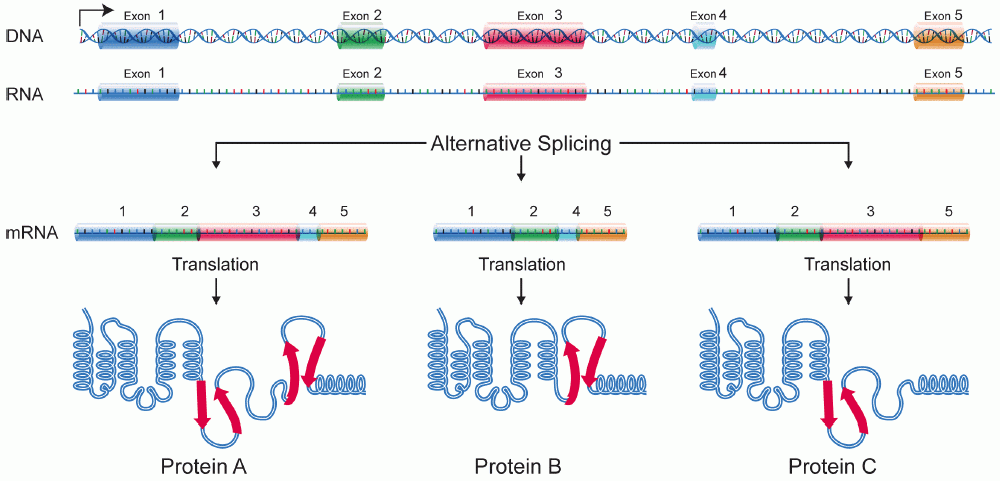
[In this image] An example of alternative splicing.
This gene contains five exons. Depending on the choice of splicing sites, one gene could generate three versions of proteins. Protein A contains all five exons. Protein B splices out exon 3 while protein C leaves out exon 4.
Image credit: wiki
After these post-transcriptional modifications, a pre-RNA becomes a mature messenger RNA (mRNA). End modifications increase the stability of the mRNA, while splicing gives the mRNA its correct sequence. If the introns are not removed correctly, they’ll be translated along with the exons, producing a “mutated” protein. Several genetic diseases may be the result of splicing errors. For example, mutations that cause the incorrect splicing of β-globin mRNA are responsible for some cases of β-thalassemia.

[In this image] In 1993, Richard J. Roberts and Phillip Allen Sharp received the Nobel Prize in Physiology or Medicine for their discovery of “split genes”. They also discovered that the splicing of the mRNA could occur in different ways, opening up the possibility for a mutation to occur.
Photo credit: The Nobel Prize
The regulation of Individual genes
Not all genes are transcribed all the time. Instead, transcription is controlled spatially (in different cells) and temporarily (in different timing) for each gene. For example, some growth hormones express only when we are young. Some proteins which can wake our immune system up will only be produced when our bodies sense pathogen invasion. Cells carefully regulate transcription, just transcribing the genes whose products are needed at a particular moment.

[In this image] Diagram showing an imaginary cell’s RNAs at a given moment.
In this cell, genes 1, 2, and 3, are transcribed, while gene 4 is not. Also, genes 1, 2, and 3 are transcribed at different levels, meaning that different numbers of RNA molecules are made for each.
Image credit: Khan Academy
Like building a dam upstream of a river, our cells control gene expressions at the beginning of transcription. There are many ways to regulate the transcription:
1. The initiation of transcription for some genes requires the binding of assistant proteins, called transcription factors, together with RNA polymerases, to their promoters.
2. Some rarely used genes have their promoters covered by inhibitory proteins or hidden deeply in folded chromosomes, which make these genes inaccessible for transcription. This is called epigenetic regulation.
3. Some genes are regulated by the stability of their mRNA. Some mRNA have pretty short half-lives and break down even before the translation happens. Only when a stimulus kicks in and makes the mRNA stable can these genes function.
Transcription in bacteria
In this article, we mainly discuss the transcription in eukaryotic cells. Prokaryotes produce RNA in a very similar way. However, there are some differences between prokaryotic and eukaryotic gene expression.
In bacteria, all transcription is performed by a single type of RNA polymerase. Because bacteria have no nucleus, their transcription and translation can occur simultaneously in the cell’s cytoplasm.
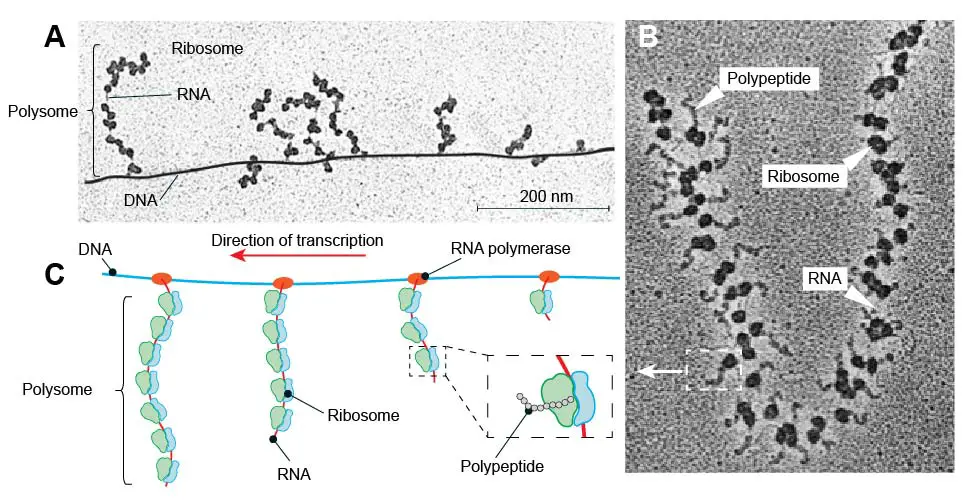
[In this figure] Bacterial transcription and translation at the same time result in the formation of “polysomes”.
Some bacterial genes group together and under the control of the same promoter. This cluster of coregulated genes is called an operon. Operon allows all its gene turned on or off together, which benefit the bacteria to rapidly adapt to changes in the environment. A bacterial mRNA encoding several genes from an operon is called polycistronic mRNA.
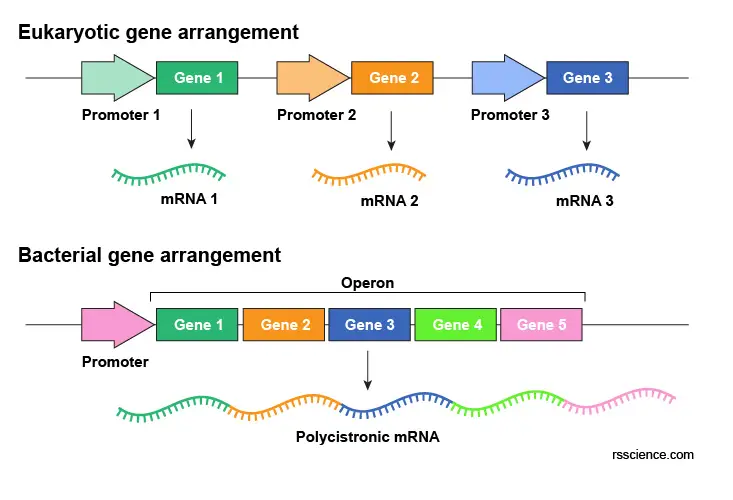
[In this figure] The comparison of eukaryotic and bacterial gene arrangement.
Each eukaryotic gene is controlled independently under its own promoter. Some bacterial genes cluster together to form an operon and are under the control of the same promoter.
The difference between prokaryotic and eukaryotic gene expression
| Bacterial gene expression | Eukaryotic gene expression |
| Transcription and translation occur simultaneously in the cytoplasm. | Transcription occurs inside the nucleus and translation occurs in the cytoplasm. |
| Has a single type of RNA polymerase. | Has three types of RNA polymerases. |
| Several functionally-related genes occur in an operon. | Individual genes are regulated separately. |
| There is no intron interrupting a gene. | Eukaryotic genes divide into introns and exons. |
| Does not include post-transcriptional modifications | Include post-transcriptional modifications (5’ cap, poly-A tail, and splicing) |
Summary
1. Transcription copies a gene’s DNA sequence to make an RNA molecule. Transcription is the first step of gene expression.
2. Transcription is done in the cell nucleus by an enzyme called RNA polymerase. RNA polymerase uses a DNA strand as a template to make an RNA copy.
3. Transcription can be divided into three stages: (1) initiation, (2) elongation, and (3) termination.
4. Transcription starts from the promoter region of a gene. Transcription initiation involves the collaboration of several proteins, including transcription factors and RNA polymerase.
5. In eukaryotes, RNA molecules must be processed after transcription – they are spliced and added 5′ caps and poly-A tails on their ends.
6. After transcription, messenger RNA (mRNA) molecules will be transported from the cell nucleus to the ribosomes in the cytoplasm, where the mRNA directs the synthesis of the protein.
7. Transcription is controlled separately for each gene in your genome. The spatially and temporally controlled expressions of different genes shape the behaviors of our cells.